Abstract
A mechanism for the coupled translocation of substrate and H+ by the lactose permease of Escherichia coli is proposed, based on a variety of experimental observations. The permease is composed of 12 α-helical rods that traverse the membrane with the N and C termini on the cytoplasmic face. Four residues are irreplaceable with respect to coupling, and the residues are paired—Arg-302 (helix IX) with Glu-325 (helix X) and His-322 (helix X) with Glu-269 (helix VIII). In an adjacent region of the molecule at the interface between helices VIII and V is the substrate translocation pathway. Because of this arrangement, interfacial changes between helices VIII and V are transmitted to the interface between helices IX and X and vice versa. Upon ligand binding, a structural change at the interface between helices V and VIII disrupts the interaction between Glu-269 and His-322, Glu-269 displaces Glu-325 from Arg-302, and Glu-325 is protonated. Simultaneously, protonated Glu-325 becomes inaccessible to water, which drastically increases its pKa. In this configuration, the permease undergoes a freely reversible conformational change that corresponds to translocation of the ternary complex. To return to ground state after release of substrate, the Arg-302–Glu-325 interaction must be reestablished, which necessitates loss of H+ from Glu-325. The H+ is released into a water-filled crevice between helices IX and X which becomes transiently accessible to both sides of the membrane due to a change in helix tilt, where it is acted upon equally by either the membrane potential or the pH gradient across the membrane.
Keywords: bioenergetics, symport, membrane proteins, charge pairs, conformational change
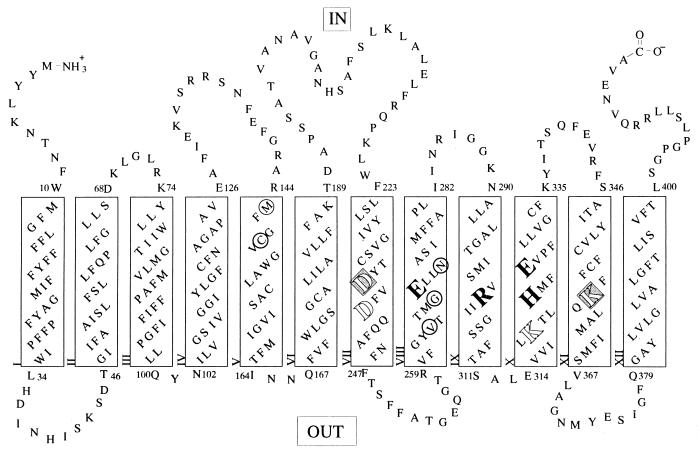
The lactose permease (lac permease) of Escherichia coli is a polytopic integral membrane protein encoded by the lacY gene and is a paradigm for proteins that transduce free energy stored in electrochemical ion gradients into solute concentration gradients (i.e., secondary active transport), a fundamental problem that is unsolved mechanistically (reviewed in refs. 1 and 2).† In the presence of a H+ electrochemical gradient across the membrane (Δμ̄H+, interior negative and/or alkaline), the permease utilizes free energy released from downhill translocation of H+ to drive accumulation of β-galactosides against a concentration gradient. Importantly, in the absence of Δμ̄H+, lac permease catalyzes the converse reaction, utilizing free energy released from downhill translocation of β-galactosides to drive uphill translocation of H+ with generation of Δμ̄H+, the polarity of which depends upon the direction of the substrate concentration gradient. The permease has been solubilized from the membrane, purified, and reconstituted, and it has been found to be solely responsible for the coupled stoichiometric translocation of β-galactosides and H+ (i.e., symport) as a monomer (reviewed in refs. 3 and 4). All available experimental evidence indicates that the protein is composed of 12 α-helical rods that traverse the membrane with N and C termini on the cytoplasmic face (Fig. 1) (reviewed in ref. 5; see refs. 6–12 in addition).
Figure 1.
Secondary structure model of lac permease. The one-letter amino acid code is used, and putative transmembrane helices are shown in boxes. The irreplaceable residues [Glu-269 (helix VIII), Arg-302 (helix IX), His-322 (helix X), and Glu-325 (helix X)] are enlarged and emboldened; Asp-237 (helix VII) and Lys-358 (helix XI) are highlighted; Asp-240 (helix VII) and Lys-319 (helix X) are shown as enlarged empty letters; and the residues thought to make direct contact with substrate [Met-145 and Cys-148 (helix V), and Val-264, Gly-268, and Asn-272 (helix VIII)] are encircled.
Tertiary Structure.
Cys-scanning and site-directed mutagenesis of almost every residue in lac permease reveal that as few as four residues are irreplaceable with respect to active lactose transport—Glu-269 (helix VIII), Arg-302 (helix IX), His-322 (helix X), and Glu-325 (helix X)—and the residues are in close proximity (Fig. 2) (reviewed in ref. 13). The spatial relationships are based on site-directed excimer fluorescence, which shows that helix VIII (Glu-269) is close to helix X (His-322), helix IX (Arg-302) is close to helix X (Glu-325), and that helix X is in α-helical conformation (His-322/Glu-325). In addition, there are two pairs of interacting Asp and Lys residues [Asp-237 (helix VII) and Lys-358 (helix XI) and Asp-240 (helix VII) and Lys-319 (helix X)] that are not essential for activity, but the interactions demonstrate that helix VII is close to helices X and XI. Evidence for the Asp–Lys pairs is derived primarily from second-site suppressor analysis, site-directed mutagenesis, and chemical rescue experiments demonstrating that neutral replacements for either Asp or Lys lead to inactivation, but double neutral replacements for Asp-237 and Lys-358 or Asp-240 and Lys-319 or neutralization of the unpaired charge in the single mutants yield permease with highly significant activity (see ref. 14). Importantly, moreover, Asp-237 can be exchanged with Lys-358 without loss of activity, whereas reversal of Asp-240 and Lys-319 inactivates (15). Parenthetically, it is noteworthy that the Asp-237–Lys-358 (or D237K/K358D‡) pair is needed for optimal insertion of the permease into the membrane, indicating that the C-terminal half of the permease is inserted into the membrane post-translationally (16).
Figure 2.
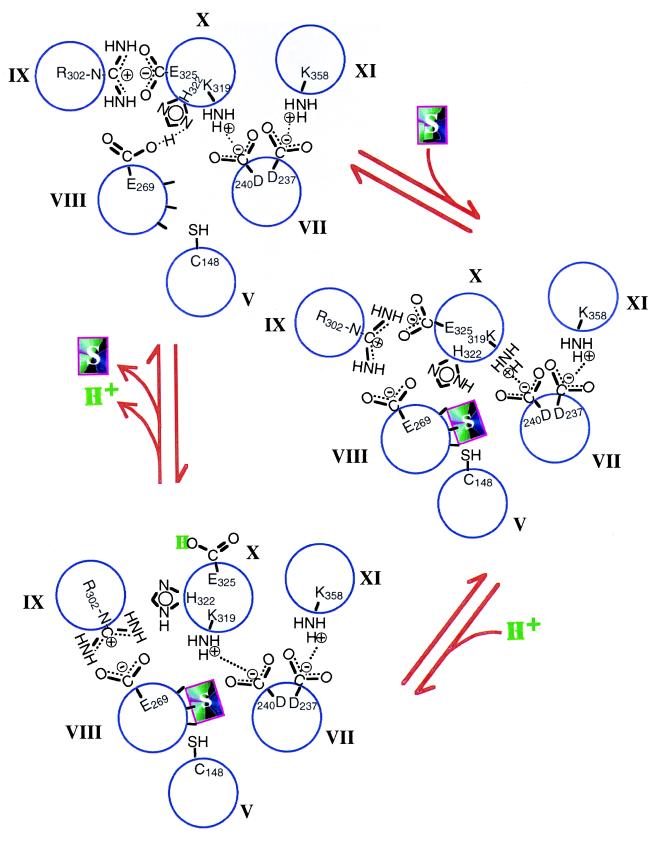
Proposed mechanism for energy coupling in lac permease. Packing of helices V and VII–XI and important side-chain interactions in the absence of substrate (Upper Left) are based on site-directed excimer fluorescence, which demonstrates that helix VIII (Glu-269) is close to helix X (His-322), helix IX (Arg-302) is close to helix X (Glu-325), and that helix X is in α-helical conformation. The presence of two pairs of charged residues that interact functionally—Asp-237 (helix VII) with Lys-358 (helix XI) and Asp-240 (helix VII) with Lys-319 (helix X)—demonstrates that helix VII is close to helices X and XI. The spatial relationships have been confirmed by engineering divalent metal-binding sites (bis-His residues) within the permease. In addition, mAb 4B11 has been shown to bind to the last two cytoplasmic loops. Site-directed chemical cleavage studies support the positioning of helix X next to helices VII and XI and indicate further that helix V is in close proximity to helices VII and VIII. The relationship between helices V, VII, and VIII has been confirmed by site-directed spin labeling and thiol crosslinking experiments. Upon substrate binding (Lower Right), the interaction between His-322 and Glu-269 is disrupted, leading to the changes in side-chain interactions described in the text. Although not shown, site-directed thiol crosslinking studies demonstrate that helix I is close to helices V and VI, helix II is close to helices VII and XI, and helix VI is close to helices V and VIII.
Many of the spatial relationships have been confirmed by engineering divalent metal-binding sites (bis-His residues) within the permease. These studies demonstrate that permease with bis-His residues at positions 269 and 322, 302 and 325, or 237 and 358 (but not at 240 and 319) binds Mn2+ with a stoichiometry of unity, a Kd in the μM range, and an apparent pKa of about 6.3. Therefore, although these positions are presumably buried in the membrane, the sites are readily accessible to water. Site-directed chemical cleavage confirms the positioning of helix X next to helices VII and XI and indicates further that helix V is close to helices VII and VIII (17). The relationship between helices V, VII, and VIII has been documented further by site-directed spin labeling and thiol crosslinking experiments (18).
Independent support for close proximity between helices VIII to XI is provided by the demonstration (11) that monoclonal antibody (mAb) 4B11 binds to an epitope composed of the last two cytoplasmic loops in the permease. In addition, the helix packing model is consistent with a number of distance measurements between an engineered Cu2+ binding site and spin-labeled single-Cys residues in this region of the permease (J. Voss, W. L. Hubbell, and H.R.K., unpublished results; see refs. 8 and 19). Although not shown in Fig. 2, recent site-directed thiol crosslinking studies demonstrate that helix I is close to helices V and VII, helix II is close to helices VII and XI (20), and helix VI is close to helices V and VIII (J. Wu and H.R.K., unpublished observations).
Properties of Mutants in the Irreplaceable Residues.
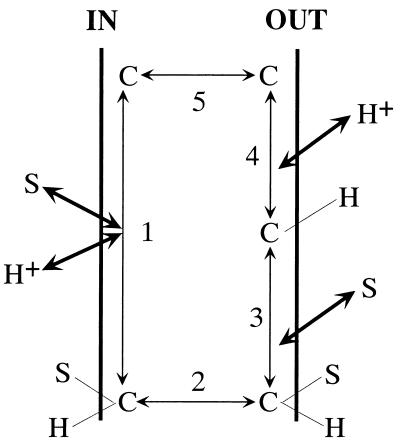
Differences in the transport properties of mutants indicate that Glu-269, Arg-302, His-322, and Glu-325 are critical for the coupling between substrate and H+ translocation. Because individual steps in the overall translocation cycle cannot be delineated by studying Δμ̄H+-driven active transport, carrier-mediated efflux down a chemical gradient, equilibrium exchange, and entrance counterflow are used to probe the mechanism (21, 22). Efflux, exchange and counterflow with wild-type permease are explained by a simple kinetic scheme (Fig. 3). Efflux consists of five steps: 1, binding of lactose and H+ to the permease at the inner surface of the membrane (order unspecified); 2, a conformational change in the permease that results in translocation of lactose and H+ to the outer surface of the membrane; 3, release of substrate; 4, release of H+; and 5, a conformational change corresponding to return of the unloaded permease to the inner surface of the membrane. Alternatively, exchange and counterflow involve steps 1–3 only. Efflux, exchange, and counterflow are blocked in His-322 and Arg-302 mutants, although the mutants catalyze lactose influx down a concentration gradient. In contrast, Glu-325 mutants are specifically defective in all steps involving net H+ translocation but catalyze exchange and counterflow as well or better than wild-type (reviewed in ref. 23). Thus, Glu-325 is required for step 4. Furthermore, Glu-325 mutations are mimicked by 2H2O or mAb 4B1, and affinity is unaffected by neutral replacements, 2H2O, or mAb 4B1. Replacement of Arg-302 with Lys or His-322 with Arg yields permease that does not catalyze active transport, exchange, or counterflow, making it unlikely that disruption of salt bridges between the irreplaceable residues per se leads fortuitously to the properties described.
Figure 3.
Schematic representation of reactions involved in lactose efflux, exchange. and counterflow. C represents lac permease; S is substrate (lactose). The order of substrate and H+ binding at the inner surface of the membrane is not implied because it was not tested. From ref. 21.
Permease with Asp in place of Glu-325 exhibits about 20% of wild-type activity and is markedly defective with respect to efflux down a concentration gradient. Remarkably, equilibrium exchange is pH dependent: below pH 7.5, exchange is rapid and the rate is comparable to wild-type; above 7.5, the rate decreases and is nil at pH 9.5, with a midpoint at about 8.5; and inhibition at alkaline pH is completely reversible (24). In contrast, wild-type exchange is only mildly inhibited above pH 9.5, and exchange by E325A or E325Q permease is comparable to wild type and is unaffected by pH.
Although none of the His-322 mutants catalyze active transport, permease with Tyr or Phe in place of His-322 (25–27) exhibits sugar-dependent H+ transport with low efficiency, and melibiose efflux remains coupled to H+ translocation. In addition, the reactions involving exchange are limiting for lactose but not melibiose efflux, and a double mutant with Val in place of Ala-177 and Asn in place of His-322 catalyzes lac-dependent H+ influx with a stoichiometry close to unity (28). Therefore, although His-322 is irreplaceable with respect to active transport, it does not appear to play a direct role in H+ translocation. Similarly, mutants in Arg-302 catalyze little or no lactose accumulation, but mutant R302S exhibits lactose-dependent H+ influx (29, 30).
Mutants with Asp, Gln, or Cys in place of Glu-269 do not catalyze active transport of lactose, equilibrium exchange, counterflow, facilitated influx, or efflux down a concentration gradient (31). Remarkably, however, E269D permease accumulates the high-affinity analog β-d-galactopyranosyl 1-thio-β-d-galactopyranoside (thiodigalactoside; TDG) in a partially uncoupled fashion with an increase in H+/TDG stoichiometry, and with the exception of galactose (S. Frillingos and H.R.K., unpublished observations), other substrates tested are not accumulated. These findings and others (32) indicate that Glu-269, like His-322, plays an essential role in the mechanism, but is not directly involved in H+ translocation.
mAb 4B1 Alters the pKa of a Carboxylic Acid at Position 325.
mAb 4B1 binds to an epitope (Phe-247, Phe-250, and Gly-254) in the periplasmic domain between helices VII and VIII (Fig. 1), uncoupling lactose from H+ translocation in much the same manner as Glu-325 mutants or 2H2O (i.e., only those reactions involving net H+ translocation are inhibited with little or no effect on translocation of the ternary complex; Fig. 3) (see ref. 10). Since no residue in this domain is important for transport and avidin binding to a biotinylated Cys residue in the domain has no effect on activity, it was suggested that 4B1 binding exerts a torsional effect which perturbs helix VII and/or VIII and alters the pKa(s) of a residue(s) critical for coupling. Consistent with this idea, 4B1 binding shifts the midpoint of the pH profile for exchange by mutant E325D from pH 8.5 to pH 7.5 (24). Since the midpoint presumably reflects the pKa of Asp-325 and mAb binding is unaffected at the pH values tested, 4B1 may cause an acid shift in the pKa of Asp-325.
The Coupling Mechanism.
The findings lead to a model for coupling that is based upon two propositions: (i) Glu-325 is the only residue in lac permease that is directly involved in H+ translocation; and (ii) in the absence of substrate, the permease does not catalyze H+ translocation.
The six helices at the heart of the mechanism form an inverted triangle with helix V at the apex, helices IX, X, and XI at the base and helices VII and VIII at the sides (Fig. 2). Cys-148, which makes direct contact with the galactosyl moiety of the substrate, is at the top of helix V on the same face as Met-145 (not shown), which also interacts with substrate, and on the adjoining face of helix VIII are three positions (Val-264, Gly-268, and Asn-272) where the reactivity of single-Cys replacements with N-ethylmaleimide is blocked by ligand. Thus, at least part of the substrate translocation pathway lies between helices V and VIII. At the upper left-hand corner is helix IX with Arg-302, which interacts electrostatically with Glu-325 (helix X). Moving counterclockwise from Glu-325 are His-322, which is hydrogen-bonded to Glu-269 (helix VIII), and Lys-319, which interacts weakly with Asp-240 (helix VII). Finally, Asp-237 (helix VII) forms a stable salt bridge with Lys-358 (helix XI). As a result of these interactions, substrate-induced structural changes at the interface between helices V and VIII are transmitted through the network of interacting residues to the interface between helices IX and X. Conversely, changes between helices IX and X are transmitted to the interface between helices V and VIII.
The postulated mechanism is as follows (Fig. 2): (i) Ligand binding at the interface between helices V and VIII induces a conformational change that disrupts the interaction between His-322 (helix X) and Glu-269 (helix VIII). (ii) Glu-269 then displaces Glu-325 (helix X) from Arg-302 (helix IX). (iii) These changes associated with substrate binding brings Glu-325 into a nonpolar environment, which increases the pKa markedly and further weakens the interaction between Lys-319 (helix X) and Asp-240 (helix VII). In this configuration, the permease catalyzes equilibrium exchange and counterflow. (iv) The total energy of the ternary complex is greater than that of the ground state. However, to return to ground state after release of ligand, the electrostatic interaction between Arg-302 and Glu-325, as well as the interaction between His-322 and Glu-269, must be reestablished, which necessitates deprotonation of Glu-325. Since bis-His residues at positions 302 and 325 form a divalent metal binding site with an apparent pKa that approximates an unperturbed imidazole, it seems reasonable to assume that there is a water-filled crevice between helices IX and X. Thus, when the H+ is released from Glu-325 between helices IX and X, it is acted upon equally by either the membrane potential or the pH gradient across the membrane.
In addition to providing a rationale for coupling between H+ and substrate translocation and an explanation for why the electrical potential and the pH gradient have the same kinetic as well as thermodynamic effect on transport, the model explains other important observations: (i) Although Asp-237 and Lys-358 can be reversed without adversely effecting permease activity, reversal of Asp-240 and Lys-319 inactivates. Clearly, according to the postulated mechanism, placement of a carboxylate at position 319 will compete with Glu-269 for His-322. On the other hand, reversal of Asp-237 and Lys-358 has no effect on activity because neither residue is sufficiently close to influence the residues directly involved in coupling or H+ translocation. (ii) According to the proposed mechanism, neither His-322 nor Glu-269 is directly involved in H+ translocation, but both residues play important roles in the coupled structural changes postulated. In addition, both residues lie close to the interface between helices V and VIII, where substrate binding is postulated to occur, and their interaction may act to stabilize this interface. (iii) As discussed above, when Glu-325 is replaced with Asp, the permease is partially uncoupled, and equilibrium exchange becomes pH dependent. Activity is normal up to pH 7.5 and decreases sharply between pH 7.5 and 9.5, with a midpoint at about pH 8.5. The simplest interpretation is that the conformational change corresponding to the translocation of the ternary complex (Fig. 3, step 2) does not tolerate a charge at position 325. In wild-type permease, the interaction between Arg-302 and Glu-325 is broken upon ligand binding, and the carboxylate is protonated and becomes inaccessible to solvent by coming into contact with the hydrophobic phase of the membrane (Fig. 2). With Asp at position 325, the side chain is less accessible to the hydrophobic phase of the membrane and more accessible to water, conditions which act to decrease the pKa relative to Glu-325. By increasing the solvent accessibility of Asp-325 in the E325D mutant, mAb 4B1 would decrease the pKa of the carboxylic acid at this position. In contrast, the pKa of a Glu residue at position 325 (i.e., the wild-type) would not be perturbed as much by 4B1 because the side chain protrudes further into the hydrophobic phase of the membrane. (iv) Recent experiments (33, 34) demonstrate that a Cys residue in place of Thr-265 (helix VIII), which lies between Glu-269 and Asn-272, Gly-268 and Val-264, residues that probably make direct contact with substrate, undergoes a marked increase in accessibility to water upon ligand binding. The finding strongly supports the idea that substrate binding induces a conformational change at the interface between helices V and VIII.
One aspect of the mechanism that is not readily visualized in Fig. 2 is the ability of the permease to catalyze H+/lactose symport in both directions across the membrane. In this respect, although the H+/lactose stoichiometry is 1:1 for influx, it is likely that the stoichiometry is less than unity for efflux. Thus, when membrane vesicles are loaded with lactose and diluted 200-fold, a membrane potential of only about −40 mV is generated versus a value of about −135 mV which is expected if the stoichiometry approximates unity (21). Although the pH gradient generated under these conditions was not measured, it is unlikely to account for the magnitude of the discrepancy. Thus, the permease may function more efficiently in one direction than the other. In any case, the type of rigid-body movement that takes place between the helices during turnover may involve changes in tilt which result in reciprocal opening and closing of partial pathways (i.e., crevices) between helices, thereby allowing for the movement of substrate and H+ in either direction across the membrane. In this respect it is noteworthy that Val-264, Gly-268, and Asn-272 are on one face of helix VIII in the periplasmic half and distributed over two turns (Fig. 1), which is greater than the length of a disaccharide. Furthermore, Cys-148 and Met-145, which also make direct contact with substrate, are on one face of helix V near the cytoplasmic face of the membrane, and Cys-148 is accessible from both sides of the membrane. The findings are more consistent with a pathway than with a fixed binding site.
One important aspect of any working model is whether it can be tested. If three-dimensional structures of the permease were available in the presence and absence of ligand, a number of questions could be resolved, but all attempts to obtain high-resolution crystals of the permease have failed thus far. On the other hand, the model makes certain explicit predictions that can be tested by means of the site-directed techniques that have been developed. One of the principal ideas in the model is that Arg-302 can interact with either Glu-325 or Glu-269, and it has been demonstrated (M. M. He, J. Voss, W. L. Hubbell, and H.R.K, unpublished results) that R302H/E269H permease binds Mn2+ with properties similar to those of R302H/E325H permease. In addition, recent experiments (Q. Wang, K. Matsushita, M. LeMaire, B. de Foresta and H.R.K., unpublished results) with permease mutants containing single-Trp residues in helix X and bromododecyl maltoside as a collisional quenching agent provide a preliminary indication that helix X moves in the presence of ligand in such a manner that the face with Glu-325 comes into closer contact with the hydrophobic phase of the membrane. By studying substrate protection of single-Cys-148 permease against labeling with a fluorescent maleimide (35), it has been demonstrated (M. M. He and H.R.K., unpublished results) that Arg-302 or Glu-325 mutants bind thiodigalactoside, lactose, and galactose normally, while Glu-269 or His-322 mutants are markedly defective. The findings are consistent with the concept that, in addition to their involvement in coupling, the interaction between Glu-269 and His-322 may stabilize the interface between helices VIII and V, the site of substrate translocation. Finally, chemical modification studies (J. Voss, J. Sun, and H.R.K., unpublished results) provide further evidence that a negative charge at position 325 can determine the rate of translocation of the ternary complex (Fig. 3, step 2). Mutant E325C catalyzes equilibrium exchange, and peroxide-catalyzed oxidation of the Cys residue to a sulfinic or sulfonic acid yields an apparent pKa of about 6.5–7.0. As discussed above, E325D permease exhibits an apparent pKa for exchange of about 8.5. Since the pKa of sulfinic or sulfonic acid is lower than that of aspartic acid, the finding supports the argument that a negative charge at position 325 is prohibitive with respect to translocation of the ternary complex.
The heart of the mechanism is thought to involve only 6 of the 12 helices in the permease. However, N-ethylmaleimide inactivates approximately 10% of the single-Cys replacement mutants in the permease, and the positions of these mutants cluster on helical faces in a manner indicating that surface interactions between the helices are important for turnover (reviewed in ref. 13; see refs. 33 and 36 in addition). It has also been demonstrated by site-directed fluorescence and labeling experiments with radioactive N-ethylmaleimide that ligand binding results in widespread conformational changes. Furthermore, crosslinking helices VII and II, neither of which contains an irreplaceable residue, inactivates the permease (37). Therefore, it is suggested that rigid body movements of the helices initiated at the interface between helices V and VIII are transmitted cooperatively throughout the molecule in order for turnover to occur.
In conclusion, this communication provides a data-based model for coupling substrate and H+ translocation in lac permease that is consistent with currently available data and makes a number of predictions which are now being tested experimentally.
Acknowledgments
I am particularly indebted to Bernard Babior for time and effort spent thinking about the proposed mechanism when he should have been writing a grant proposal. I also express my deep appreciation to the members of my laboratory and to David Sigman for many spirited discussions and for critical reading of the manuscript and to Kerstin Stempel for preparing the figures. Some of the experiments described were supported in part by National Institutes of Health Grant 1 R0 1 DK51131–01.
ABBREVIATIONS
- lac permease
lactose permease
- Δμ̄H+
the H+ electrochemical gradient across the membrane
Footnotes
To limit the reference list to a reasonable size, review articles are referenced rather than original research articles whenever possible.
Site-directed mutants are designated as follows: the one-letter amino acid code is used followed by a number indicating the position of the residue in the wild-type lac permease, and then a second letter denoting the amino acid replacement at this position.
References
- 1.Kaback H R. J Cell Physiol. 1976;89:575–593. doi: 10.1002/jcp.1040890414. [DOI] [PubMed] [Google Scholar]
- 2.Kaback H R. J Membr Biol. 1983;76:95–112. doi: 10.1007/BF02000610. [DOI] [PubMed] [Google Scholar]
- 3.Viitanen P, Newman M J, Foster D L, Wilson T H, Kaback H R. Methods Enzymol. 1986;125:429–452. doi: 10.1016/s0076-6879(86)25034-x. [DOI] [PubMed] [Google Scholar]
- 4.Sahin-Tóth M, Lawrence M C, Kaback H R. Proc Natl Acad Sci USA. 1994;91:5421–5425. doi: 10.1073/pnas.91.12.5421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaback H R. In: International Review of Cytology. Friedlander M, Mueckler M, editors. 137A. New York: Academic; 1992. pp. 97–125. [DOI] [PubMed] [Google Scholar]
- 6.Zen K H, McKenna E, Bibi E, Hardy D, Kaback H R. Biochemistry. 1994;33:8198–8206. doi: 10.1021/bi00193a005. [DOI] [PubMed] [Google Scholar]
- 7.Zen K, Consler T G, Kaback H R. Biochemistry. 1995;34:3430–3437. doi: 10.1021/bi00010a035. [DOI] [PubMed] [Google Scholar]
- 8.Voss J, Hubbell W L, Kaback H R. Proc Natl Acad Sci USA. 1995;92:12300–12303. doi: 10.1073/pnas.92.26.12300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Voss J, He M, Hubbell W, Kaback H R. Biochemistry. 1996;35:12915–12918. doi: 10.1021/bi9608774. [DOI] [PubMed] [Google Scholar]
- 10.Sun J, Wu J, Carrasco N, Kaback H R. Biochemistry. 1996;35:990–998. doi: 10.1021/bi952166w. [DOI] [PubMed] [Google Scholar]
- 11.Sun J, Li J, Carrasco N, Kaback H R. Biochemistry. 1997;36:274–280. doi: 10.1021/bi962292f. [DOI] [PubMed] [Google Scholar]
- 12.Lee J I, Varela M F, Wilson T H. Biochim Biophys Acta. 1996;1278:111–118. doi: 10.1016/0005-2736(95)00209-x. [DOI] [PubMed] [Google Scholar]
- 13.Kaback H R. In: Handbook of Biological Physics: Transport Processes in Eukaryotic and Prokaryotic Organisms. Konings W N, Kaback H R, Lolkema J S, editors. Vol. 2. Amsterdam: Elsevier; 1996. pp. 203–227. [Google Scholar]
- 14.Frillingos S, Kaback H R. Biochemistry. 1996;35:13363–13367. doi: 10.1021/bi961453c. [DOI] [PubMed] [Google Scholar]
- 15.Sahin-Tóth M, Dunten R L, Gonzalez A, Kaback H R. Proc Natl Acad Sci USA. 1992;89:10547–10551. doi: 10.1073/pnas.89.21.10547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dunten R L, Sahin-Tóth M, Kaback H R. Biochemistry. 1993;32:3139–3145. doi: 10.1021/bi00063a028. [DOI] [PubMed] [Google Scholar]
- 17.Wu J, Perrin D, Sigman D, Kaback H. Proc Natl Acad Sci USA. 1995;92:9186–9190. doi: 10.1073/pnas.92.20.9186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu J, Voss J, Hubbell W L, Kaback H R. Proc Natl Acad Sci USA. 1996;93:10123–10127. doi: 10.1073/pnas.93.19.10123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Voss J, Salwinski L, Kaback H R, Hubbell W L. Proc Nat Acad Sci USA. 1995;92:12295–12299. doi: 10.1073/pnas.92.26.12295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu J, Kaback H R. Proc Natl Acad Sci USA. 1996;93:14498–14502. doi: 10.1073/pnas.93.25.14498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaczorowski G J, Kaback H R. Biochemistry. 1979;18:3691–3697. doi: 10.1021/bi00584a009. [DOI] [PubMed] [Google Scholar]
- 22.Kaczorowski G J, Robertson D E, Kaback H R. Biochemistry. 1979;18:3697–3704. doi: 10.1021/bi00584a010. [DOI] [PubMed] [Google Scholar]
- 23.Kaback H R. Biochemistry. 1987;26:2071–2076. doi: 10.1021/bi00382a001. [DOI] [PubMed] [Google Scholar]
- 24.Frillingos S, Kaback H R. Biochemistry. 1996;35:10166–10171. doi: 10.1021/bi960995r. [DOI] [PubMed] [Google Scholar]
- 25.King S C, Wilson T H. J Biol Chem. 1989;264:7390–7394. [PubMed] [Google Scholar]
- 26.King S C, Wilson T H. Biochim Biophys Acta. 1989;982:253–264. doi: 10.1016/0005-2736(89)90062-x. [DOI] [PubMed] [Google Scholar]
- 27.King S C, Wilson T H. J Biol Chem. 1990;265:3153–3160. [PubMed] [Google Scholar]
- 28.Brooker R J. J Biol Chem. 1990;265:4155–4160. [PubMed] [Google Scholar]
- 29.Menick D R, Carrasco N, Antes L M, Patel L, Kaback H R. Biochemistry. 1987;26:6638–6644. doi: 10.1021/bi00395a012. [DOI] [PubMed] [Google Scholar]
- 30.Matzke E A, Stephenson L J, Brooker R J. J Biol Chem. 1992;267:19095–19100. [PubMed] [Google Scholar]
- 31.Ujwal M L, Sahin-Tóth M, Persson B, Kaback H R. Mol Membr Biol. 1994;1:9–16. doi: 10.3109/09687689409161024. [DOI] [PubMed] [Google Scholar]
- 32.Franco P J, Brooker R J. J Biol Chem. 1994;269:7379–7386. [PubMed] [Google Scholar]
- 33.Frillingos S, Ujwal M L, Sun J, Kaback H R. Protein Sci. 1997;6:431–437. doi: 10.1002/pro.5560060220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frillingos S, Kaback H R. Protein Sci. 1997;6:438–443. doi: 10.1002/pro.5560060221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu J, Kaback H R. Biochemistry. 1994;33:12166–12171. doi: 10.1021/bi00206a020. [DOI] [PubMed] [Google Scholar]
- 36.Frillingos S, Sun J, Gonzalez A, Kaback H R. Biochemistry. 1997;36:269–273. doi: 10.1021/bi9618629. [DOI] [PubMed] [Google Scholar]
- 37.Wu, J. & Kabac, H. R. (1997) J. Mol. Biol., in press.