Abstract
Study Objectives:
The synchronization of peripheral circadian oscillators in humans living on atypical sleep/wake schedules is largely unknown. In this night shift work simulation, we evaluate clock gene expression in peripheral blood mononuclear cells (PBMCs) relative to reliable markers of the central circadian pacemaker.
Design:
Participants were placed on a 10-hr delayed sleep/wake schedule simulating nighttime work followed by a daytime sleep episode.
Setting:
Baseline, intermediate and final circadian evaluations were performed in the temporal isolation laboratory.
Participants:
Five healthy candidates, 18-30 years.
Interventions:
Polychromatic white light of (mean ±SEM) 6,036 ±326 lux (∼17,685 ±955 W/m2) during night shifts; dim light exposure after each night shift; an 8-hr sleep/darkness episode beginning 2 hrs after the end of each night shift.
Measurements:
Melatonin and cortisol in plasma; clock genes HPER1, HPER2 and HBMAL1 RNA in PBMCs.
Results:
Following 9 days on the night schedule, hormonal rhythms were adapted to the shifted schedule. HPER1 and HPER2 expression in PBMCs displayed significant circadian rhythmicity, which was in a conventional relationship with the shifted sleep/wake schedule. Changes in the pattern of clock gene expression were apparent as of 3 days on the shifted sleep/wake schedule.
Conclusions:
This preliminary study is the first documentation of the effects of a shifted sleep/wake schedule on the circadian expression of both peripheral circadian oscillators in PBMCs and centrally-driven hormonal rhythms. In light of evidence associating clock gene expression with tissue function, the study of peripheral circadian oscillators has important implications for understanding medical disorders affecting night shift workers.
Citation:
James FO; Cermakian N; Boivin DB. Circadian rhythms of melatonin, cortisol, and clock gene expression during simulated night shift work.. SLEEP 2007;30(11):1427-1436.
Keywords: Night shift work, melatonin, cortisol, HPER1, HPER2, HBMAL1
INTRODUCTION
NIGHT SHIFT WORK HAS BEEN ASSOCIATED WITH A NUMBER OF NEGATIVE HEALTH OUTCOMES INCLUDING CARDIOVASCULAR AND GASTROINTESTINAL disease,1 disturbances of the metabolic syndrome,2 reproductive difficulties including spontaneous abortion,3 and increased risk of cancer.4 The rapid reorientation in the sleep/wake schedule brought about by night shift work results in a misalignment between the output of the endogenous circadian pacemaker and the shifted schedule that generally persists despite consecutive shifts worked.5 Light is the primary synchronizer of the endogenous circadian pacemaker of the suprachiasmatic nucleus of the anterior hypothalamus (SCN), and careful control over light and darkness exposure throughout the day can promote a more appropriate temporal alignment between the circadian pacemaker and the sleep/wake schedule.6–9
The central circadian pacemaker of the SCN is comprised of neurons that demonstrate an intrinsic rhythmicity in their electrical and metabolic activity.10 The characteristic activity of the SCN is dependant on the expression of auto-regulatory loops of so-called circadian clock genes including Clock, Bmal1, three Period genes (Per 1, 2, 3), and two cryptochrome genes (Cry 1, 2), such that individual clock genes demonstrate a circadian rhythm in the levels of transcripts in SCN neurons.11 It has been demonstrated that the rhythmic expression of circadian clock genes required for the intrinsic rhythmicity of the SCN is also present in non-SCN tissues.11–13 Clock gene expression in peripheral tissues is particularly of interest since it has been associated with elements of tissue function such as cytokine and cytolytic factor production by NK cells,14,15 B cell maturation,16 adipogenesis,17 fatty acid metabolism in cardiomyocytes,18 bone formation,19 glucocorticoid synthesis,20 and regulation of the cell-cycle.21 However, despite the relative independence of peripheral clock function, it is currently held that the SCN coordinates the expression of cellular rhythmicity across peripheral tissues.13,22–24 Evidence for the presence of functional peripheral circadian oscillators in humans has been recently reported. Rhythmic clock gene expression has been observed in human oral mucosa and skin samples25 and in peripheral blood mononuclear cells (PBMCs).11,26–29 Under constant conditions, the expression of core clock genes HPER1 and HPER2 displays a significant circadian oscillation in PBMCs and peaks a few hours after that of plasma melatonin concentration.28
The aim of this simulated shift work experiment was to evaluate the temporal relationship between clock gene expression in PBMCs and the shifted sleep/wake schedule in humans. In transgenic rats, the pattern of Per1-driven luminescence in the SCN rapidly demonstrates complete adaptation to shifts in the light/darkness schedule such that the expected time of peak Per1 expression occurs in its habitual temporal relationship with the shifted light/darkness schedule.12 However, the entrainment of peripheral oscillators in skeletal muscle or the liver tissue, for example, does not necessarily occur as rapidly as in the SCN.12,30 In order to investigate the response of human peripheral oscillators to shifted schedules, this experiment took advantage of an intervention based on a pattern of light and darkness exposure known to efficiently entrain centrally-driven circadian rhythms, such as plasma melatonin and cortisol, to night shifts.
MATERIALS AND METHODS
Subjects
Five candidates, 4 male and 1 female, all nonsmoking and drug-free (group mean age ± SD: 24.9 ± 4.8 yr) with normal BMI (23.4 ± 1.6 kg/m2) were certified to be in good mental and physical health and free of intrinsic sleep disorders following medical and psychological interviews. Each candidate gave informed consent to their participation in this study approved by the Douglas Mental Health University Institute Research Ethics Board and within the ethical standards of the Declaration of Helsinki. Selected candidates had no history of night shift work or travel across time zones in the three months preceding the study. The female participant had regular menses with an average cycle duration of 32 ± 1 days. She entered the laboratory on the second day after the start of menses and the entire investigation took place within the follicular phase of her menstrual cycle. All experiments were performed in the months of July and August 2005.
Experimental Protocol
For 2 weeks before the start of the investigation, participants maintained stable sleep schedules including a single 8-hr nighttime sleep episode and restricting naps. The prestudy sleep schedule was based on participants′ reported habitual sleep times and durations. For all participants, habitual sleep durations reported during the recruitment phase ranged between 8 and 9 hrs. The start and end of the sleep/darkness episode during the prestudy period were within (±SD) 0.4 ±0.5 hr and 0.6 ±0.6 hr of reported bed and wake times, respectively. The maintenance of a stable sleep/darkness schedule serves to reduce the variability in the expression of circadian markers.8 Participants recorded times in and out of bed in sleep/wake logs and left voice messages with the laboratory to confirm these times. Sleep/wake schedules were also verified using wrist actigraphy. Mean prestudy (baseline) reported times in bed are shown in Table 1. During the prestudy period, mean sleep/darkness period lengths were within 10 min of the 8-hr target sleep/darkness period.
Table 1.
Mean Prestudy Reported Times in Bed
| Baseline |
Night shift schedule |
|||
|---|---|---|---|---|
| Time in bed | Time out of bed | Time in bed | Time out of bed | |
| T01 | 23:25 | 7:30 | 9:30 | 17:28 |
| T02 | 22:01 | 6:04 | 8:05 | 16:05 |
| T04 | 00:00 | 8:01 | 10:02 | 18:06 |
| T05 | 23:26 | 7:29 | 9:29 | 17:36 |
| T06 | 23:58 | 8:08 | – | – |
Mean times in and out of bed reported in sleep/wake logs for each participant. During the baseline (pre-study) period, subjects maintained a habitual day-oriented sleep/wake schedule prior to their admission to the laboratory. Each maintained an 8-hr sleep/darkness period that was based on their reported habitual bed and wake times. During the ambulatory phase of the investigation (experimental days 6-10), participants continued to maintain the night-oriented schedule by remaining awake at night and sleeping during the day while at home. Subject T06 did not participate in the last segment of the investigation (experimental days 6-12).
The experimental protocol for the 12-day investigation is shown in Figure 1. The first 6 days of the experiment took place in the time-free laboratory environment. After one night on their habitual sleep/wake schedules, subjects underwent a 9-day simulated night shift work procedure. The sleep/wake schedule was delayed by 10 hrs relative to habitual sleep/wake times and was maintained for the remainder of the study. From the evening of experimental day 6 to the evening of experimental day 10, subjects maintained the experimental conditions while at home. Subjects were readmitted to the laboratory on experimental day 10.
Figure 1.
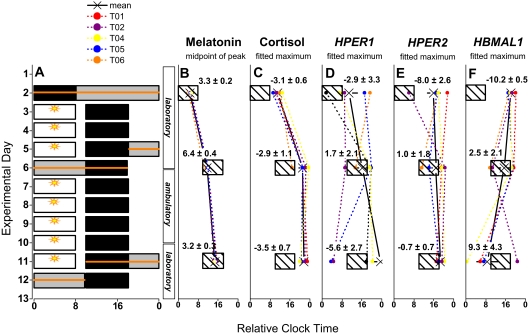
Experimental protocol and mean shifts in circadian markers. A: The 12-day experimental protocol is described in the Methods section. Successive experimental days are shown along the vertical axis and time is shown on the horizontal axis. Scheduled sleep/darkness periods are shown as black rectangles. For illustrative purposes, bedtimes for nighttime and daytime sleep episodes were assigned relative clock times of 00:00 and 10:00, respectively. Periods of bright light exposure during simulated night shifts both in the laboratory and during the ambulatory phase of the study are shown as open bars with sun symbols. The expression of markers of the endogenous circadian pacemaker and of clock gene in PBMCs were measured from 24-hr blood sampling sessions performed before the start of simulated night shifts (experimental day 2), after three days on the shifted schedule (experimental days 5-6), and at the end of the shifted schedule (experimental days 11-12). Blood sampling periods are shown as horizontal lines overlaid on the protocol. B-F: For each individual, the mean phase of each marker is shown relative to night or daytime sleep/darkness periods (hatched boxes representing night and day sleep episodes). Group mean phase angles are shown in decimal hrs ±SEM and demonstrate the relationship between the sleep/wake schedule and circadian phase. One subject did not participate in the final evaluation, thus results of this evaluation are given for n=4 subjects, while n=5 for the first two evaluations.
On the day of laboratory admission, participants remained in ordinary indoor room light via ceiling-mounted banks of cool-white fluorescent fixtures covered with lenses emitting less than 1% radiant energy up to 400nm (4,100K Philips, Somerset, NJ, USA, and Sylvania, Danvers, MA, USA). In the laboratory, light intensity was measured in the angle of gaze with a research photometer (IL1400A, International Light, Newburyport, MA, USA). Given irradiance values were subsequently approximated for radiance of 400-700 nm from cool white fluorescent bulbs. Mean intensity in the angle of gaze ±SD, was 144 ±62 lux (∼422 ±182 W/m2) during laboratory admission on experimental day 1. On experimental day 2 and thereafter, ambient light levels during periods of wakefulness remained dim (6 ±3 lux; (∼18 ±9 W/m2) except when subjects were exposed to bright, polychromatic white light during their 8-hr simulated night shifts. Participants wore long sleeves and pants throughout the phototherapy period. During the period of bright light exposure, subjects were seated and were asked to center their gaze on a point on a wall for 10 of every 20 min (the “up” period).31 The angle of gaze was left unrestricted for the remaining 10 min of the 20-min lapse (the “down” period). Mean levels of light exposure measured in the angle of gaze during “up” periods were 6,036 ±728 lux; (∼17,685 ±2,133 W/m2). Mean light intensity during down periods was approximately 80% of mean intensities measured during “up” periods. Participants slept in darkness (∼0.03 lux; < 0.1 W/m2).
During the ambulatory segment of the investigation, polychromatic white light was provided by portable lamps equipped with fluorescent bulbs covered with an ultraviolet filter throughout each 8-hr simulated night shift (Sunsquare 23″×24″, 5,000 K, Sunbox Company, Gaithersburg, MD, USA; maximum light intensity measured with a research photometer at 50 cm is ∼10,000 lux, ∼29,300 W/m2). During simulated night shifts, participants were asked to remain seated before the lamp at a designated work table for as much of the 8-hr period as possible. Mean levels of light exposure during phototherapy performed both in the laboratory and at home were 1,213 ±555 lux (∼ 3,554 ±1,626 W/m2) as measured via wrist-mounted actigraphy (Actiwatch-L, Mini-Mitter, Bend, OR). During simulated night shifts at home, participants left messages with the laboratory voicemail to confirm their compliance with the ambulatory protocol every 30 min. Participants slept in their own bedrooms, darkened by research staff, and reduced their exposure to morning light in the 2 hrs preceding sleep/darkness by wearing sunglasses with 5% visual light transmission (neutral gray filter, North, Tornado, T57005GRYM, The Litebook Company, Medicine Hat, AB). Participants were also asked to wear their sunglasses when outdoors during evening hours before sunset. The maintenance of sleep/darkness schedules was verified by sleep/wake logs, regular telephone calls to the laboratory and wrist-actigraphy monitoring. During this ambulatory segment, participants reported sleep/darkness periods within 10 min of the 8-hr target period length (Table 1).
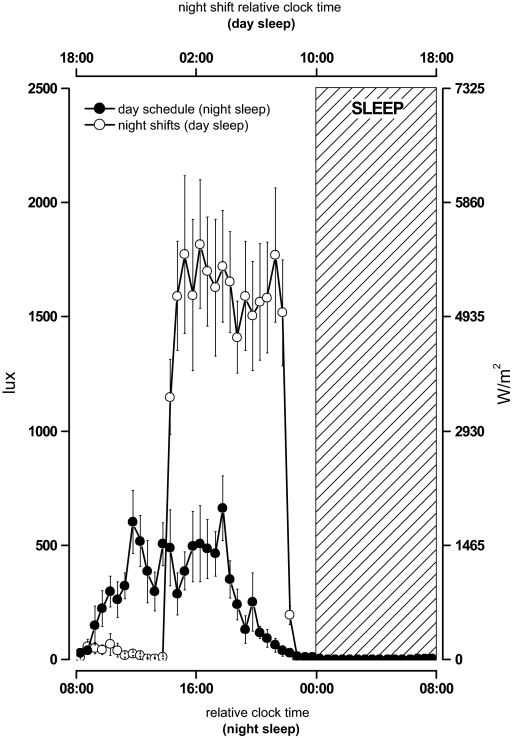
Light data was sampled throughout the investigation—including the prestudy period—via wrist-mounted actigraphy with light sensor. The 24-hr pattern of light exposure for the group of participants is shown in Figure 2. For every day of sampling, data from each individual were aligned to the end of the sleep/darkness period and averaged into 30-min bins. Binned data were then averaged across subjects. The mean pattern of light exposure on a day-oriented schedule was based on data collected during the prestudy period and the first day in the laboratory. Actigraphy light sensor data collected both during the laboratory and ambulatory segments of the study were included in means created for the night work schedule.
Figure 2.
Pattern of light exposure during pre-study period and simulated night shifts: Light intensity data (in lux) was collected via wrist-mounted actigraphy with light sensor during the laboratory and ambulatory segments of the investigation. Irradiance values, approximated for radiation of 400-700 nm from cool-white fluorescent bulbs, are given along the right y-axis. For each individual, data were aligned relative to the end of the sleep/darkness period and averaged into 30-min bins before creating a group mean (shown here as ±SEM). On the lower x-axis, the start of the sleep/darkness episode is assigned a relative clock time of midnight. Light intensity data collected while participants maintained a day-oriented schedule are shown as closed symbols. Light intensity data collected while participants maintained the shifted sleep/wake schedule are shown as open symbols. Data were corrected for use of sunglasses as reported by participants in experimental logs during the ambulatory segment of the study. On the upper x-axis, the initiation of the daytime sleep/darkness episode is assigned a relative clock time of 10:00. The 8-hr sleep/darkness episode is shown as a hatched rectangle.
Meal times were standardized during the prestudy period and throughout the study, including the ambulatory segment. Subjects took breakfast 30 min after awakening, lunch 5 hrs after awakening, dinner 11 hrs after awakening, and an evening snack 13.5 hrs after awakening.
Circadian Phase Assessment
The expression of markers of the endogenous circadian pacemaker and of clock genes in PBMCs were measured by 24-hr blood sampling sessions performed in the laboratory before the start of simulated night shifts (experimental day 2), after 3 days on the shifted schedule (experimental days 5-6), and at the end of the shifted schedule (experimental days 11-12). Each sampling period included a 16-hr constant posture (CP) period of limited activity where participants maintained a semi-recumbent posture in bed. Meals were replaced by hourly, nutritionally balanced snacks.
Circadian phase assessment for markers of the central pacemaker (plasma cortisol and melatonin) and peripheral oscillators (clock gene expression in PBMCs) were performed by repeated hourly blood sampling via an indwelling catheter. Circadian phase of the plasma melatonin rhythm was defined from hourly whole blood samples as the midpoint between the upward and downward crossing of the 24-hr average of plasma melatonin concentration.6 The phase of the cortisol rhythm was defined as the time of the fitted maximum of cortisol concentration, based on a single harmonic regression applied to hourly sampling data.7
Peripheral blood mononuclear cells were isolated from whole blood samples drawn every ∼120 min on a density gradient (Histopaque-1077, Sigma-Aldrich Canada, Oakville, ON, Canada) and stored at −80°C in Trizol reagent (Invitrogen Canada, Burlington, ON, Canada). RNA was extracted and reverse transcribed using MultiScribe Reverse Transcriptase (Applied Biosystems, Foster City, CA, USA). Quantification of clock gene expression was performed by real-time PCR using SYBR Green chemistry (Applied Biosystems, Foster City, CA, USA). The expression of clock genes HPER1, HPER2, HBMAL1 was described relative to the expression of HCDK4 28 using the following primers:
HPER1
Forward: 5′-TGGCTATCCACAAGAAGATTC-3′
Reverse: 5′-GGTCAAAGGGCTGGCCCG-3′
HPER2
Forward: 5′-GGCCATCCACAAAAAGATCCTGC-3′
Reverse: 5′-GAAACCGAATGGGAGAATAGTCG-3′
HBMAL1
Forward: 5′-GGCTCATAGATGCAAAAACTGG-3′
Reverse: 5′-CTCCAGAACATAATCGAGATGG-3′
HCDK4
Forward: 5′-ATCCCAATGTTGTCCGGCTG-3′
Reverse: 5′-TGATCTCCCGGTCAGTTCGG-3′
Fold change in gene expression of each clock gene for each sample was determined using the 2-ΔΔCT calculation to quantify levels of gene expression relative to the internal control, and then expressed as a proportion of the maximum for each 24-hr blood sampling evaluation. A statistically significant circadian oscillation was observed where the 95% confidence interval for the amplitude of the expression estimated by a dual-harmonic regression analysis with a period search of 23-26 hr32 did not include the zero value.28 The amplitude of expression was defined as the mean-to-trough difference of the first harmonic of the regression.33 All data points collected were included in the regression analyses performed for each individual. The phase of clock gene expression in PBMCs was defined as the time of fitted maximum of expression.
To demonstrate the relationship between the sleep/wake schedule and circadian phase in central and peripheral markers, phase angles were calculated as: (end of scheduled sleep/darkness period) – (circadian phase) for each individual. Thus, positive and negative phase angles indicate that the measured phase of the circadian marker occurred before and after the end of the scheduled sleep/darkness period, respectively.
For each laboratory evaluation, we also determined a profile for the expression of each circadian marker across a total of 6 wake or sleep bins corresponding to the 24-hr day. Individual data were first aligned to the habitual time of the end of the planned sleep/dark period, and each data point was assigned to a 4-hr bin representing Wake (W1-W4) or Sleep (S1-S2) periods throughout the 24-hr day, data were averaged per bin per subject, and then averaged across all subjects. The width of the analysis bin was selected to provide the greatest resolution while permitting individuals to equally contribute to each analysis bin despite occasional missed samples. Two-factor ANOVA for repeated measures (time-of-day and sampling session) were performed on binned values.
Results are expressed as mean ± SEM.
RESULTS
The midpoint of melatonin expression occurred 3.3 ±0.2, and 3.2 ±0.3 hrs before the end of the planned sleep/dark period in the baseline and final evaluations, respectively (Figure 1). During these blood sampling sessions, the fitted maximum of cortisol occurred 3.1 ±0.6, and 3.5 ±0.7 hr after the end of the sleep/darkness period, respectively. The alignment of melatonin and cortisol rhythms with the sleep/wake cycle were comparable in the baseline and final evaluations (t-test, P = 0.7, and P = 0.9, respectively). As observed in night shift workers receiving a similar intervention,6,7 these markers displayed an appropriate alignment to the sleep/wake cycle in night and daytime sleep schedules (Figures 3 and 4).
Figure 3.
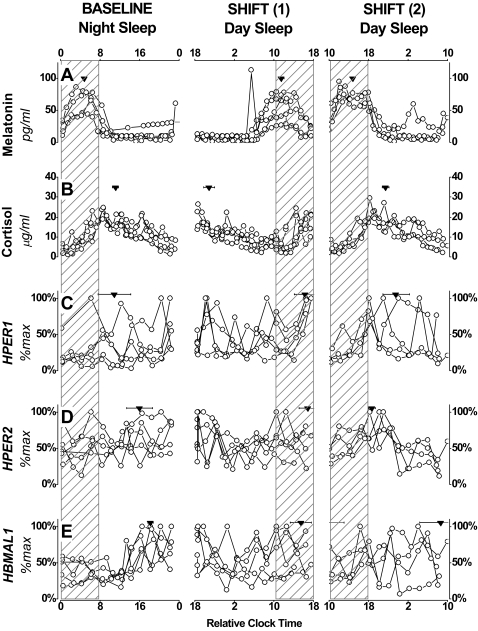
Central and peripheral clock markers relative to sleep/wake schedules. Individual data for melatonin (A), cortisol (B), HPER1 (C), HPER2 (D), HBMAL1 (E) sampled during 24-hr blood sampling sessions performed before and during the maintenance of a night shift work schedule. Group mean phase ±SEM is shown for each marker as an inverted triangle. Sleep/darkness periods are shown as hatched rectangles. For illustrative purposes, bedtimes for nighttime and daytime sleep episodes were assigned relative clock times of 00:00 and 10:00, respectively. The expression of markers of the endogenous circadian pacemaker and of clock genes in PBMCs were measured from 24-hr blood sampling sessions performed at baseline before the start of simulated night shifts (experimental day 2), after three days on the shifted schedule (shift 1; experimental days 5-6), and at the end of the shifted schedule (shift 2; experimental days 11-12).
Figure 4.
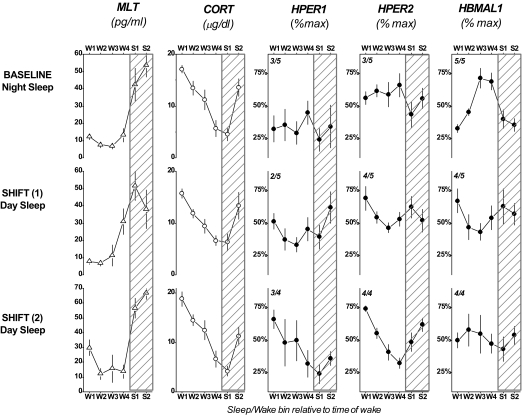
Expression of circadian markers throughout the sleep/wake cycle. Four-hr bins for Wake (W1-W4) or Sleep (S1-S2) represent the 24-hr day. For each clock gene evaluation, the fraction of individuals for whom a significant circadian amplitude of expression was detected is shown in the upper left corner of each cell. One subject did not participate in the final evaluation, thus results for this evaluation are given for n=4 subjects.
Central and peripheral clock markers are shown for each individual in Figure 3. Fitted parameters of clock gene expression for each individual are summarized in Table 2. During the baseline evaluation, the time of fitted maximum of HPER1, HPER2 and HBMAL1 occurred at 2.9 ±3.3, 8.0 ±2.6, and 10.2 ±0.5 hr after the scheduled end of the sleep/darkness period, respectively. At the end of 9 days on the shifted sleep/wake schedule, the fitted peak of HPER1 and HPER2 expression occurred 5.6 ±2.7 and 0.7 ±0.7 after the end of the sleep/darkness period, respectively. The fitted peak of HBMAL1 expression occurred 9.4 ±4.3 before (or 14.6 ±4.3 hr after) the end of the planned sleep/darkness period. No statistically significant difference was detected in the alignment of HPER1 and HPER2 maxima relative to the sleep/wake schedule between the initial and final conditions (t-test, P = 0.6 and P = 0.2, respectively), although these phase angles were more variable than those calculated for hormonal rhythms, such that a reliable phase shift could not be calculated. The alignment of HBMAL1 expression tended to differ in the initial and final conditions (P = 0.07).
Table 2.
Fitted Parameters of Clock Gene Expression for Individual Participants
| BS1 (Baseline) |
BS2 (Night shift) |
BS3 (Night shift) |
||||
|---|---|---|---|---|---|---|
| amplitude | Ψwake | amplitude | Ψwake | amplitude | Ψwake | |
| HPER1 | ||||||
| T01 | *0.298 | 6.3 | *0.132 | −1.5 | *0.170 | 0.1 |
| T02 | *0.102 | −1.0 | 0.022 | 8.6 | 0.103 | −10.6 |
| T04 | 0.063 | 0.9 | 0.174 | −1.3 | *0.232 | −2.2 |
| T05 | 0.040 | −9.2 | 0.124 | −1.8 | *0.309 | −9.6 |
| T06 | *0.336 | −11.3 | *0.218 | 4.6 | — | — |
| HPER2 | ||||||
| T01 | 0.049 | −13.5 | *0.257 | −1.1 | *0.155 | 0.9 |
| T02 | *0.212 | 1.8 | 0.083 | −2.5 | *0.261 | −1.3 |
| T04 | 0.112 | −9.8 | *0.150 | −1.4 | *0.188 | −2.4 |
| T05 | *0.178 | −9.1 | *0.130 | 3.8 | *0.121 | 0 |
| T06 | *0.227 | −9.4 | *0.183 | 6.6 | — | — |
| HBMAL1 | ||||||
| T01 | *0.105 | −11.9 | *0.274 | −1.3 | *0.174 | 12.0 |
| T02 | *0.255 | −10.7 | *0.230 | 7.2 | *0.248 | −2.9 |
| T04 | *0.286 | −9.9 | *0.295 | −0.6 | *0.272 | 17.5 |
| T05 | *0.122 | −9.2 | 0.130 | −0.8 | *0.153 | 10.8 |
| T06 | *0.284 | −9.3 | *0.215 | 8.2 | — | — |
Regression analyses were performed on individual profiles of HPER1, HPER2 and HBMAL1 to determine fitted amplitude and circadian phase (time of fitted maximum expression). The relationship between circadian phase of clock gene expression and the sleep/wake cycle is given as a phase angle (Ψwake, in hrs) calculated as: [end of the sleep/darkness period – circadian phase of clock gene expression]. Thus, negative phase angles indicate that circadian phase occurred after the end of the sleep/darkness episode. Positive phase angles indicate that circadian phase occurred before the end of the sleep/darkness episode. Three 24-hr blood sampling sessions were performed before the start of simulated night shifts (experimental day 2; BS1), after three days on the night shift work shifted schedule with daytime sleep/darkness (experimental days 5-6; BS2), and at the end of the shifted schedule with daytime sleep/darkness (experimental days 11-12; BS3). Subject T06, did not participate in the final blood sampling session. Statistically significant amplitudes (i.e., 95% confidence interval does not include zero) are indicated with an asterisk (*).
The analysis of circadian markers throughout wake and sleep periods is shown in Figure 4. ANOVA for repeated measures performed on daily rhythms for all 3 blood sampling sessions revealed a significant time of day x blood sampling session interaction for melatonin (F10,55 = 2.40, P = 0.02), and a significant effect of time of day for cortisol (F5,55 = 50.01, P <0.0001). No statistically significant effects or interactions were detected by ANOVA for HPER1. A significant time of day x blood sampling session interaction was detected for HPER2 rhythms (F10,55 = 2.99, P = 0.004) where simple main effects analyses revealed a significant effect of time of day at the final blood sampling session (P = 0.001). A significant interaction was also detected for HBMAL1 (F10,55 = 3.34, P = 0.002, respectively), where simple main effects analyses revealed a significant effect of time of day at the first blood sampling session (P = 0.0006).
DISCUSSION
This study has demonstrated clock gene expression in a conventional relationship with a shifted sleep/wake schedule following an intervention known to entrain endogenous melatonin and cortisol rhythms. The time, duration, and intensity of our light/darkness intervention were expected to promote rapid and large phase delays in the expression of the central circadian pacemaker.34 Intersubject variability in HPER1 and HPER2 expression at baseline resulted in little rhythmicity in the group mean (Figure 4). However, as of 3 days on the night shift schedule the pattern of clock gene expression began to adapt to the shifted sleep/wake schedule. By the final assessment, the alignment of clock gene expression rhythms was similar to those reported for day-active subjects (Figure 4). Specifically, group means of HPER1 and HPER2 expression demonstrated clear rhythmicity throughout the day, and both reached maximal levels in the early hrs of the scheduled wake period. This observation is in line with what has previously been described for the pattern of HPER1 and HPER2 expression in PBMCs under constant conditions and in the presence of a habitual sleep/wake schedule.27–29 Conversely, the group mean pattern of HBMAL1 expression demonstrated rhythmic changes throughout the day at baseline and was more variable at the end of the experiment although the fitted peak of HBMAL1 expression occurred late in the scheduled wake period as we would expect.11
These novel, preliminary results point out two important aspects of the expression of clock genes in peripheral blood cells: the interindividual variability and the potential adaptability to shifted schedules. The interindividual variability in phase of clock gene expression we observed, and as demonstrated in Figure 3, was present despite tightly controlled behavioral and environmental conditions. Such variability has been reported in the period32 and phase26 of clock gene expression in human peripheral oscillators, although no single factor has been identified as the source of the observed differences. Clock gene mutations and polymorphisms may contribute to diurnal preference,35 sleep regulation,36 as well as certain sleep/wake disorders.37 An observed segregation in the times of peak HBMAL1 and HPER2 expression in PBMCs has been attributed to the existence of molecular phenotypes that may in turn explain the heterogeneity in clock gene expression.26 Although none of our participants demonstrated extreme diurnal preferences, we cannot determine the extent to which these factors contributed to the variability we observed in the initial condition.
Delay in central clock markers of melatonin and cortisol were coherent with the 10-hr shift in the sleep/wake cycle. The calculation of mean phase shifts for peripheral markers based on variable initial phases would be misleading in this small group and we cannot reliably confirm the direction of phase shifts we observe. Type zero-like resetting of Bmal1 expression has been described for in vitro cultures of mouse fibroblasts following serum shock with dexamethasone.38 If this type of resetting is possible in human PBMCs in vivo, then this may explain the apparently large phase shifts observed in certain individuals as of the second blood sampling session. Moreover, anti-dromic resetting can be suspected in certain individuals. Differences between the changes in patterns of HPER1, HPER2 and HBMAL1 over the 3 blood sampling sessions may suggest that individual clock components have distinct modes for entrainment.
Disparity in the entrainment of participants′ central clock to the sleep/wake cycle at the start of the investigation is not likely to be a major contributor to the interindividual variability in clock gene expression. In the prestudy period, participants were placed on sleep/darkness schedules that were comparable to their reported habitual bedtimes. The intersubject variability in the phase angles of central clock markers was modest at the start and end of the experiment: initial and final standard deviations calculated for melatonin phase angles were both 0.5 hr, while standard deviations calculated for cortisol phase angles were 1.4 and 1.3 hr, respectively. As demonstrated in Figure S1 (available as supplementary material on www.journalsleep.org), interindividual variability in the phase of clock gene expression in PBMCs remains present when phase is replotted as a function of circadian phase of melatonin or cortisol. Nevertheless, our analysis of peripheral clock markers and their relationship to the sleep/wake schedule makes it apparent that individual patterns of HPER1 and HPER2 expression peak near the shifted time of awakening in the final condition, and are in a temporal alignment reminiscent of previously published observations,27–29 thus suggesting an entrainment to the night-oriented schedule. This strongly suggests that the experimental procedure improved the synchronization of clock gene expression to the shifted schedule, both among PBMC populations and between subjects.
There is evidence to suggest that the light/dark cycle may affect peripheral clock gene expression in human and nonhuman mammals. In rodents, the pattern of light exposure has been shown to affect the expression of clock genes in the SCN and in peripheral oscillators. However, the time required for phase shifts in extra-SCN oscillators may be longer and associated with a temporary internal desynchrony.12 In jaundiced neonates with covered eyes, blue light therapy appears to reduce mean HBMAL1 expression in PBMCs when samples collected before and after light exposure are compared.39 In healthy adults, blue light that suppresses plasma melatonin concentration was reported to induce HPER2 expression in oral mucosa samples during a 2-hr exposure.40 The specific wavelength used led the authors to suggest that the effects on HPER2 are mediated via the SCN. While a number of different tissue-specific signals may entrain peripheral oscillators,41 the mechanisms by which the SCN coordinates clock gene expression in peripheral tissues are currently under investigation. Cross-correlation analyses performed on central and peripheral markers in the present study detected no consistent and significant relationship between the resetting of cortisol or melatonin rhythms and clock gene expression in PBMCs. Interestingly, a number of experiments suggest that glucocorticoids may play a role in the communication between the SCN and peripheral oscillators in vivo.42 Indeed, HPER1 is induced in human PBMCs sampled following prednisolone therapy.43 Changes in rodent feeding schedules may shift the oscillation of peripheral clocks without shifting SCN-driven rhythms,44 and there may be an interaction between photic and nonphotic resetting factors on peripheral clock gene expression.30 We cannot determine from our experiment the specific contributions of the shifted sleep/wake schedule, the meal schedule and the schedule of light/darkness exposure on clock gene expression. It is tempting to suggest that in the presence of the light intervention, an SCN-directed entrainment signal was powerful enough to both reduce interindividual differences contributing to the variability observed in the baseline conditions and entrain clock gene expression to the shifted sleep/wake schedule. Indeed, the 24-hr patterns of light exposure in the prestudy period and during the investigation (Figure 2) confirm that participants were more often exposed to bright levels of light during the simulated night shift experiment. If the strength of the Zeitgeber to the central clock affects the expression of peripheral clocks, then the weaker light stimulus received in the prestudy period could explain the greater interindividual variability in peripheral clock markers at the start of the investigation. Moreover, the greater interindividual variability observed in peripheral clock markers compared to melatonin and cortisol phase suggests that the factors known to entrain the central clock would entrain peripheral clocks in PBMCs more weakly. Future studies will be required to partial out the nature of synchronizers of peripheral circadian oscillators, and to quantify their relative strength.
Despite the small number of individuals included in this report, the results of the present study have important practical implications for understanding the medical disorders affecting night shift workers. Namely, there exists evidence to suggest that the molecular clock may significantly affect tissue function and interact with the regulation of the cell cycle and tumorigenesis.11,21 Cancerous cells may also demonstrate abnormalities in circadian clock genes.11,45 In light of recent evidence associating increased risk of certain cancers with night shift work,11 our results demonstrate that circadian adaptation to night shift work may be advantageous on a fundamental level. As discussed, there is also evidence to indicate that functional peripheral circadian oscillators are important in the tissue function such as in the cardiovascular system.18 Thus, a crucial step in reducing the health problems associated with night shift work will be to fully understand the regulation of central and peripheral circadian oscillators in humans.
ACKNOWLEDGMENTS
We wish thank the research subjects for their participation and the research staff and students of the Laboratory of Molecular Chronobiology and the Centre for Study and Treatment of Circadian Rhythms for their invaluable contributions to this investigation. We thank Dr. France Varin of the University of Montréal for hormonal assays. We wish to express our gratitude to Ms. Doris Dea and Dr. Judes Poirier for access to real-time PCR equipment and assistance in troubleshooting assays. We are also indebted to Dr. Sylvie Rhéaume for medical supervision during the investigation and to Dr. Jean Roy for providing medical consultations. The experiments described were conducted with the approbation of the Douglas Mental Health University Institute Research Ethics board and within Canadian legal standards. We thank the members of the Research Ethics Board for their guidance in the preparation of this study.
Funding: This study was made possible by a Fonds de Recherche en Santé du Québec salary award (NC), as well as a Canadian Institutes of Health Research grant (DBB, NC) and salary award (DBB).
Figure S1.
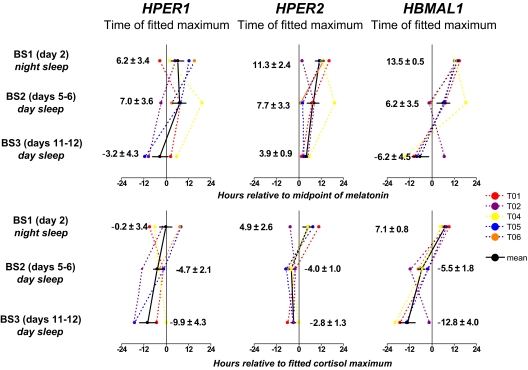
The expression of central and peripheral clock markers relative to the circadian phase of (A) melatonin and (B) cortisol. For each individual, the fitted phase HPER1, HPER2 and HBMAL1 at each of three 24-hr blood sampling sessions (BS1, BS2, BS3) is shown relative to the expression of a central clock marker (melatonin in upper panels; cortisol in lower panels). Each point represents a phase angle calculated between the phase of the central (melatonin or cortisol) and peripheral clock markers. (Phase angle = [Phaseperipheral] - [Phasecentral]). Positive values indicate that the phase of the peripheral marker occurred afterthe phase of the of the central clock marker; negative values indicate that phase of the peripheral marker occurred before that of the central marker. Means for the group are shown in each panel ± SEM. The phase of central clock marker was assigned a value of zero.
Footnotes
Disclosure Statement
This is not an industry supported study. Dr. James has consulted for Circadian Health Improvement Inc. Dr Boivin has received research support from Transport Canada and the Litebook Company; has participated in speaking and advisory engagements for Servier and Cephalon and is CEO and Scientific Director of Circadian Health Improvement, Inc. Dr. Cermakian has indicated no financial conflicts of interest.
REFERENCES
- 1.Knutsson A. Health disorders of shift workers. Occup Med (Lond) 2003;53:103–8. doi: 10.1093/occmed/kqg048. [DOI] [PubMed] [Google Scholar]
- 2.Karlsson B, Knutsson A, Lindahl B. Is there an association between shift work and having a metabolic syndrome? Results from a population based study of 27,485 people. Occup Environ Med. 2001;58:747–52. doi: 10.1136/oem.58.11.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu JL, Hjollund NH, Andersen AM, Olsen J. Shift work, job stress, and late fetal loss: The National Birth Cohort in Denmark. J Occup Environ Med. 2004;46:1144–9. doi: 10.1097/01.jom.0000145168.21614.21. [DOI] [PubMed] [Google Scholar]
- 4.Davis S, Mirick DK. Circadian disruption, shift work and the risk of cancer: a summary of the evidence and studies in Seattle. Cancer Causes Control. 2006;17:539–45. doi: 10.1007/s10552-005-9010-9. [DOI] [PubMed] [Google Scholar]
- 5.Boivin DB. Disturbances of hormonal circadian rhythms in shift workers. In: Cardinali DP, Pandi-Perumal SR, editors. Neuroendocrine correlates of sleep/wakefulness. New York: Springer; 2005. pp. 325–54. [Google Scholar]
- 6.Boivin DB, James FO. Circadian adaptation to night-shift work by judicious light and darkness exposure. J Biol Rhythms. 2002;17:556–67. doi: 10.1177/0748730402238238. [DOI] [PubMed] [Google Scholar]
- 7.James FO, Walker CD, Boivin DB. Controlled exposure to light and darkness realigns the salivary cortisol rhythm in night shift workers. Chronobiol Int. 2004;21:961–72. doi: 10.1081/cbi-200035944. [DOI] [PubMed] [Google Scholar]
- 8.Horowitz TS, Cade BE, Wolfe JM, Czeisler CA. Efficacy of bright light and sleep/darkness scheduling in alleviating circadian maladaptation to night work. Am J Physiol Endocrinol Metab. 2001;281:E384–91. doi: 10.1152/ajpendo.2001.281.2.E384. [DOI] [PubMed] [Google Scholar]
- 9.Crowley SJ, Lee C, Tseng CY, Fogg LF, Eastman CI. Combinations of bright light, scheduled dark, sunglasses, and melatonin to facilitate circadian entrainment to night shift work. J Biol Rhythms. 2003;18:513–23. doi: 10.1177/0748730403258422. [DOI] [PubMed] [Google Scholar]
- 10.Silver R, Schwartz WJ. The suprachiasmatic nucleus is a functionally heterogeneous timekeeping organ. Methods Enzymol. 2005;393:451–65. doi: 10.1016/S0076-6879(05)93022-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lamont EW, James FO, Boivin DB, Cermakian N. From circadian clock gene expression to pathologies. Sleep Med. 2007;8:547–56. doi: 10.1016/j.sleep.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 12.Yamazaki S, Numano R, Abe M, et al. Resetting central and peripheral circadian oscillators in transgenic rats. Science. 2000;288:682–5. doi: 10.1126/science.288.5466.682. [DOI] [PubMed] [Google Scholar]
- 13.Yoo SH, Yamazaki S, Lowrey PL, et al. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci U S A. 2004;101:5339–46. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu J, Mankani G, Shi X, et al. The circadian clock Period 2 gene regulates gamma interferon production of NK cells in host response to lipopolysaccharide-induced endotoxic shock. Infect Immun. 2006;74:4750–6. doi: 10.1128/IAI.00287-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arjona A, Sarkar DK. Evidence supporting a circadian control of natural killer cell function. Brain Behav Immun. 2006;20:469–76. doi: 10.1016/j.bbi.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 16.Sun Y, Yang Z, Niu Z, et al. MOP3, a component of the molecular clock, regulates the development of B cells. Immunology. 2006;119:451–60. doi: 10.1111/j.1365-2567.2006.02456.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shimba S, Ishii N, Ohta Y, et al. Brain and muscle Arnt-like protein-1 (BMAL1), a component of the molecular clock, regulates adipogenesis. Proc Natl Acad Sci U S A. 2005;102:12071–6. doi: 10.1073/pnas.0502383102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Durgan DJ, Trexler NA, Egbejimi O, et al. The circadian clock within the cardiomyocyte is essential for responsiveness of the heart to fatty acids. J Biol Chem. 2006;281:24254–69. doi: 10.1074/jbc.M601704200. [DOI] [PubMed] [Google Scholar]
- 19.Fu L, Patel MS, Bradley A, Wagner EF, Karsenty G. The molecular clock mediates leptin-regulated bone formation. Cell. 2005;122:803–15. doi: 10.1016/j.cell.2005.06.028. [DOI] [PubMed] [Google Scholar]
- 20.Oster H, Damerow S, Kiessling S, et al. The circadian rhythm of glucocorticoids is regulated by a gating mechanism residing in the adrenal cortical clock. Cell Metab. 2006;4:163–73. doi: 10.1016/j.cmet.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 21.Granda TG, Liu XH, Smaaland R, et al. Circadian regulation of cell cycle and apoptosis proteins in mouse bone marrow and tumor. FASEB J. 2005;19:304–6. doi: 10.1096/fj.04-2665fje. [DOI] [PubMed] [Google Scholar]
- 22.Guo H, Brewer JM, Lehman MN, Bittman EL. Suprachiasmatic regulation of circadian rhythms of gene expression in hamster peripheral organs: effects of transplanting the pacemaker. J Neurosci. 2006;26:6406–12. doi: 10.1523/JNEUROSCI.4676-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sakamoto K, Nagase T, Fukui H, et al. Multitissue circadian expression of rat period homolog (rPer2) mRNA is governed by the mammalian circadian clock, the suprachiasmatic nucleus in the brain. J Biol Chem. 1998;273:27039–42. doi: 10.1074/jbc.273.42.27039. [DOI] [PubMed] [Google Scholar]
- 24.Pando MP, Morse D, Cermakian N, Sassone-Corsi P. Phenotypic rescue of a peripheral clock genetic defect via SCN hierarchical dominance. Cell. 2002;110:107–17. doi: 10.1016/s0092-8674(02)00803-6. [DOI] [PubMed] [Google Scholar]
- 25.Bjarnason GA, Jordan RC, Wood PA, et al. Circadian expression of clock genes in human oral mucosa and skin: association with specific cell-cycle phases. Am J Pathol. 2001;158:1793–801. doi: 10.1016/S0002-9440(10)64135-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Teboul M, Barrat-Petit MA, Li XM, et al. Atypical patterns of circadian clock gene expression in human peripheral blood mononuclear cells. J Mol Med. 2005;83:693–9. doi: 10.1007/s00109-005-0697-6. [DOI] [PubMed] [Google Scholar]
- 27.Kusanagi H, Mishima K, Satoh K, Echizenya M, Katoh T, Shimizu T. Similar profiles in human period1 gene expression in peripheral mononuclear and polymorphonuclear cells. Neurosci Lett. 2004;365:124–7. doi: 10.1016/j.neulet.2004.04.065. [DOI] [PubMed] [Google Scholar]
- 28.Boivin DB, James FO, Wu A, Cho-Park PF, Xiong H, Sun ZS. Circadian clock genes oscillate in human peripheral blood mononuclear cells. Blood. 2003;102:4143–5. doi: 10.1182/blood-2003-03-0779. [DOI] [PubMed] [Google Scholar]
- 29.Takata M, Burioka N, Ohdo S, et al. Daily expression of mRNAs for the mammalian Clock genes Per2 and clock in mouse suprachiasmatic nuclei and liver and human peripheral blood mononuclear cells. Jpn J Pharmacol. 2002;90:263–9. doi: 10.1254/jjp.90.263. [DOI] [PubMed] [Google Scholar]
- 30.Hara R, Wan K, Wakamatsu H, et al. Restricted feeding entrains liver clock without participation of the suprachiasmatic nucleus. Genes Cells. 2001;6:269–78. doi: 10.1046/j.1365-2443.2001.00419.x. [DOI] [PubMed] [Google Scholar]
- 31.Boivin DB, Duffy JF, Kronauer RE, Czeisler CA. Sensitivity of the human circadian pacemaker to moderately bright light. J Biol Rhythms. 1994;9:315–31. doi: 10.1177/074873049400900311. [DOI] [PubMed] [Google Scholar]
- 32.Brown SA, Fleury-Olela F, Nagoshi E, et al. The period length of fibroblast circadian gene expression varies widely among human individuals. PLoS Biol. 2005;3:e338. doi: 10.1371/journal.pbio.0030338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jewett ME, Kronauer RE, Czeisler CA. Phase-amplitude resetting of the human circadian pacemaker via bright light: a further analysis. J Biol Rhythms. 1994;9:295–314. doi: 10.1177/074873049400900310. [DOI] [PubMed] [Google Scholar]
- 34.Duffy JF, Wright KP., Jr. Entrainment of the human circadian system by light. J Biol Rhythms. 2005;20:326–38. doi: 10.1177/0748730405277983. [DOI] [PubMed] [Google Scholar]
- 35.Carpen JD, von Schantz M, Smits M, Skene DJ, Archer SN. A silent polymorphism in the PER1 gene associates with extreme diurnal preference in humans. J Hum Genet. 2006;51:1122–5. doi: 10.1007/s10038-006-0060-y. [DOI] [PubMed] [Google Scholar]
- 36.Viola AU, Archer SN, James LM, et al. PER3 polymorphism predicts sleep structure and waking performance. Curr Biol. 2007;17:613–8. doi: 10.1016/j.cub.2007.01.073. [DOI] [PubMed] [Google Scholar]
- 37.Toh KL, Jones CR, He Y, et al. An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science. 2001;291:1040–3. doi: 10.1126/science.1057499. [DOI] [PubMed] [Google Scholar]
- 38.Nagoshi E, Saini C, Bauer C, Laroche T, Naef F, Schibler U. Circadian gene expression in individual fibroblasts: cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell. 2004;119:693–705. doi: 10.1016/j.cell.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 39.Chen A, Du L, Xu Y, Chen L, Wu Y. The effect of blue light exposure on the expression of circadian genes: bmal1 and cryptochrome 1 in peripheral blood mononuclear cells of jaundiced neonates. Pediatr Res. 2005;58:1180–4. doi: 10.1203/01.pdr.0000183663.98446.05. [DOI] [PubMed] [Google Scholar]
- 40.Cajochen C, Jud C, Munch M, Kobialka S, Wirz-Justice A, Albrecht U. Evening exposure to blue light stimulates the expression of the clock gene PER2 in humans. Eur J Neurosci. 2006;23:1082–6. doi: 10.1111/j.1460-9568.2006.04613.x. [DOI] [PubMed] [Google Scholar]
- 41.Hirota T, Fukada Y. Resetting mechanism of central and peripheral circadian clocks in mammals. Zoolog Sci. 2004;21:359–68. doi: 10.2108/zsj.21.359. [DOI] [PubMed] [Google Scholar]
- 42.Yamamoto T, Nakahata Y, Tanaka M, et al. Acute physical stress elevates mouse period1 mRNA expression in mouse peripheral tissues via a glucocorticoid-responsive element. J Biol Chem. 2005;280:42036–43. doi: 10.1074/jbc.M509600200. [DOI] [PubMed] [Google Scholar]
- 43.Fukuoka Y, Burioka N, Takata M, et al. Glucocorticoid administration increases hPer1 mRNA levels in human peripheral blood mononuclear cells in vitro or in vivo. J Biol Rhythms. 2005;20:550–3. doi: 10.1177/0748730405279866. [DOI] [PubMed] [Google Scholar]
- 44.Damiola F, Le Minh N, Preitner N, Kornmann B, Fleury-Olela F, Schibler U. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000;14:2950–61. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang MY, Chang JG, Lin PM, et al. Downregulation of circadian clock genes in chronic myeloid leukemia: Alternative methylation pattern of hPER3. Cancer Sci. 2006;97:1298–307. doi: 10.1111/j.1349-7006.2006.00331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]