Abstract
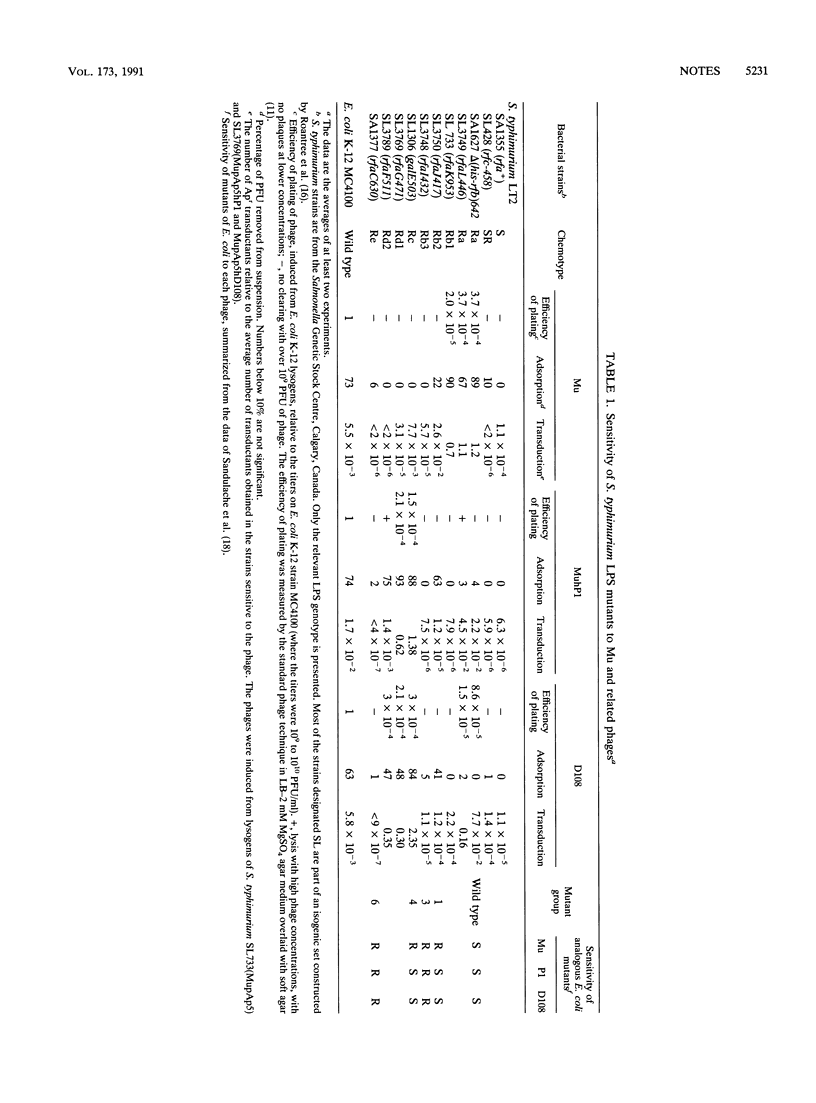
The lipopolysaccharide receptors for the mutator bacteriophages Mu, MuhP1, and D108 were investigated with lipopolysaccharide mutants of Salmonella typhimurium LT2. Mu adsorbed only to mutants lacking the terminal O antigen but retaining the main chain sugars of the core; the side chain N-acetylglucosamine was not required. MuhP1 and D108 adsorbed partially to cells with the same receptors but adsorbed well only to cells with shorter lipopolysaccharides of the Rc and Rd1 chemotypes.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bullas L. R., Ryu J. I. Salmonella typhimurium LT2 strains which are r- m+ for all three chromosomally located systems of DNA restriction and modification. J Bacteriol. 1983 Oct;156(1):471–474. doi: 10.1128/jb.156.1.471-474.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csonka L. N., Howe M. M., Ingraham J. L., Pierson L. S., 3rd, Turnbough C. L., Jr Infection of Salmonella typhimurium with coliphage Mu d1 (Apr lac): construction of pyr::lac gene fusions. J Bacteriol. 1981 Jan;145(1):299–305. doi: 10.1128/jb.145.1.299-305.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faelen M., Mergeay M., Gerits J., Toussaint A., Lefèbvre N. Genetic mapping of a mutation conferring sensitivity to bacteriophage Mu in Salmonella typhimurium LT2. J Bacteriol. 1981 Jun;146(3):914–919. doi: 10.1128/jb.146.3.914-919.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groisman E. A., Casadaban M. J. Cloning of genes from members of the family Enterobacteriaceae with mini-Mu bacteriophage containing plasmid replicons. J Bacteriol. 1987 Feb;169(2):687–693. doi: 10.1128/jb.169.2.687-693.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansson P. E., Lindberg A. A., Lindberg B., Wollin R. Structural studies on the hexose region of the core in lipopolysaccharides from Enterobacteriaceae. Eur J Biochem. 1981 Apr;115(3):571–577. doi: 10.1111/j.1432-1033.1981.tb06241.x. [DOI] [PubMed] [Google Scholar]
- Kamp D., Kardas E., Ritthaler W., Sandulache R., Schmucker R., Stern B. Comparative analysis of invertible DNA in phage genomes. Cold Spring Harb Symp Quant Biol. 1984;49:301–311. doi: 10.1101/sqb.1984.049.01.036. [DOI] [PubMed] [Google Scholar]
- Murooka Y., Takizawa N., Harada T. Introduction of bacteriophage Mu into bacteria of various genera and intergeneric gene transfer by RP4::Mu. J Bacteriol. 1981 Jan;145(1):358–368. doi: 10.1128/jb.145.1.358-368.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller K. H., Trust T. J., Kay W. W. Unmasking of bacteriophage Mu lipopolysaccharide receptors in Salmonella enteritidis confers sensitivity to Mu and permits Mu mutagenesis. J Bacteriol. 1988 Mar;170(3):1076–1081. doi: 10.1128/jb.170.3.1076-1081.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roantree R. J., Kuo T. T., MacPhee D. G. The effect of defined lipopolysaccharide core defects upon antibiotic resistances of Salmonella typhimurium. J Gen Microbiol. 1977 Dec;103(2):223–234. doi: 10.1099/00221287-103-2-223. [DOI] [PubMed] [Google Scholar]
- Roncero C., Darzins A., Casadaban M. J. Pseudomonas aeruginosa transposable bacteriophages D3112 and B3 require pili and surface growth for adsorption. J Bacteriol. 1990 Apr;172(4):1899–1904. doi: 10.1128/jb.172.4.1899-1904.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandulache R., Prehm P., Kamp D. Cell wall receptor for bacteriophage Mu G(+). J Bacteriol. 1984 Oct;160(1):299–303. doi: 10.1128/jb.160.1.299-303.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toussaint A., Lefebvre N., Scott J. R., Cowan J. A., de Bruijn F., Bukhari A. I. Relationships between temperate phages Mu and P1. Virology. 1978 Aug;89(1):146–161. doi: 10.1016/0042-6822(78)90048-x. [DOI] [PubMed] [Google Scholar]


