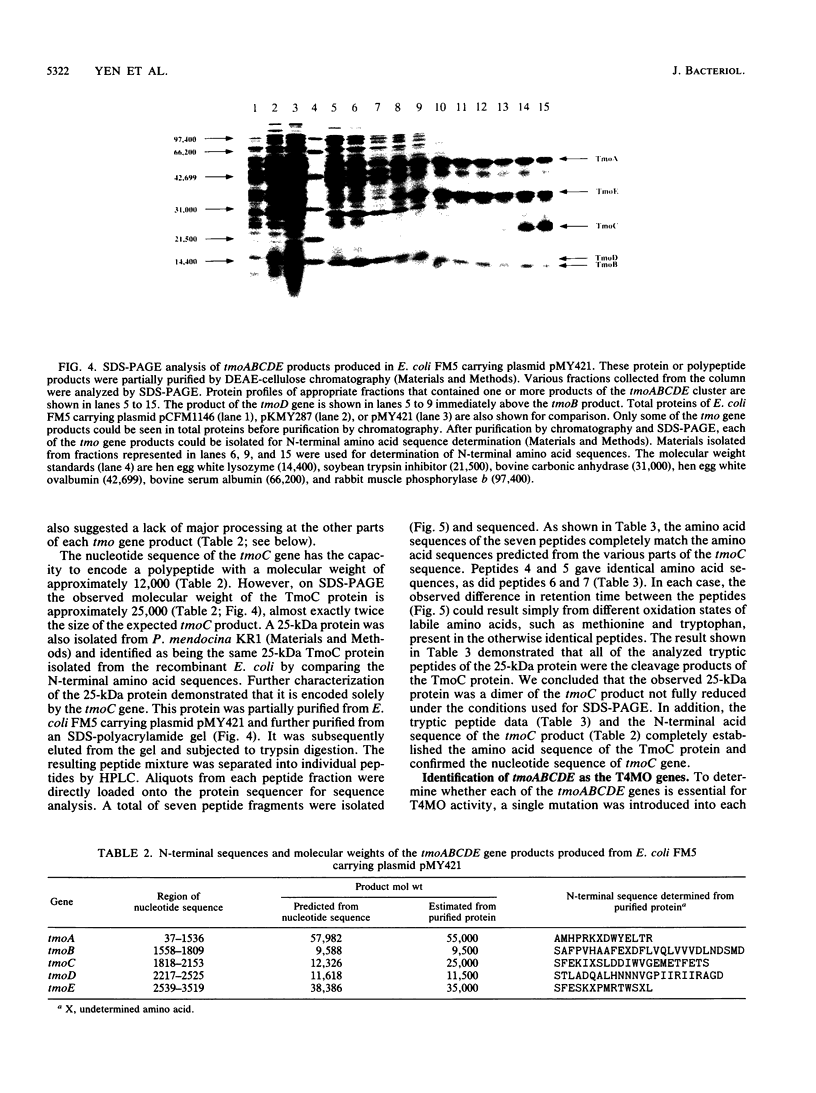
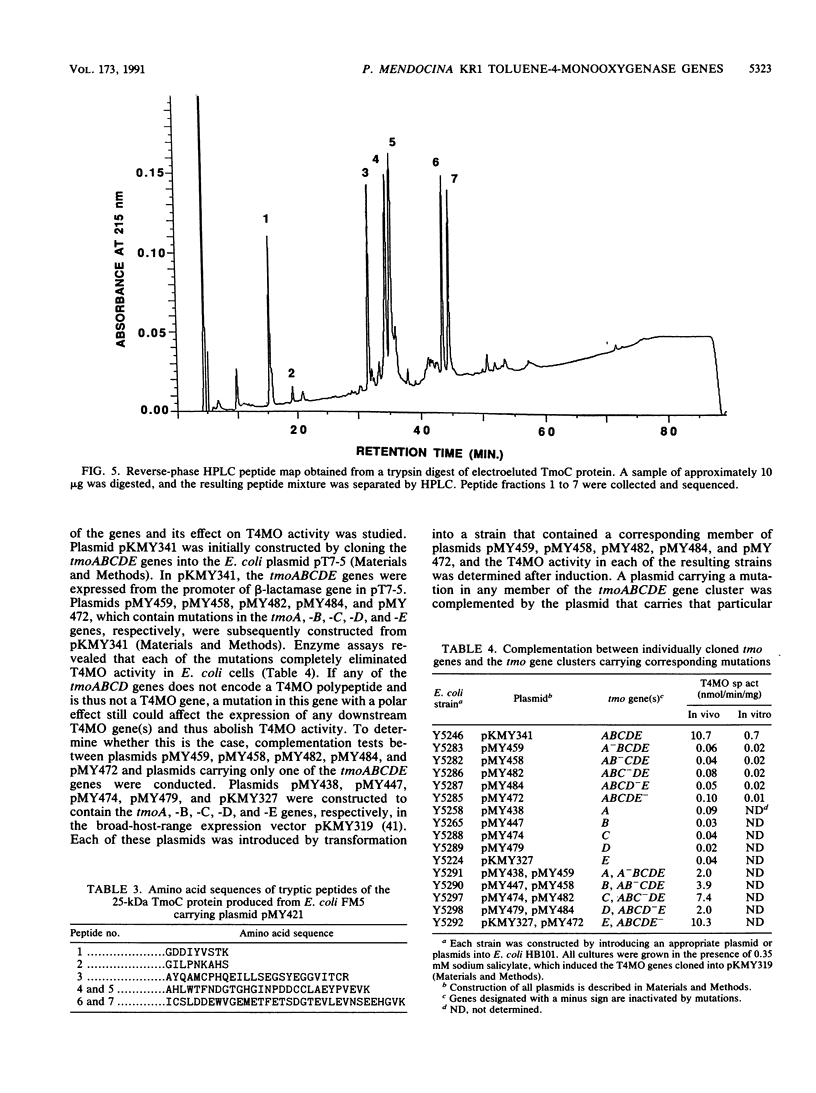
Abstract
Pseudomonas mendocina KR1 metabolizes toluene as a carbon source by a previously unknown pathway. The initial step of the pathway is hydroxylation of toluene to form p-cresol by a multicomponent toluene-4-monooxygenase (T4MO) system. The T4MO enzyme system has broad substrate specificity and provides a new opportunity for biodegradation of toxic compounds and bioconversions. Its known activities include conversion of a variety of phenyl compounds into the phenolic derivatives and the complete degradation of trichloroethylene. We have cloned and characterized a gene cluster from KR1 that determines the offO activity. To clone the T4MO genes, KR1 DNA libraries were constructed in Escherichia coli HB101 by using a broad-host-range vector and transferred to a KR1 mutant able to grow on p-cresol but not on toluene. An insert consisting of two SacI fragments of identical size (10.2 kb) was shown to complement the mutant for growth on toluene. One of the SacI fragments, when cloned into the E. coli vector pUC19, was found to direct the synthesis of indigo dye. The indigo-forming property was correlated with the presence of T4MO activity. The T4MO genes were mapped to a 3.6-kb region, and the direction of transcription was determined. DNA sequencing and N-terminal amino acid determination identified a five-gene cluster, tmoABCDE, within this region. Expression of this cluster carrying a single mutation in each gene demonstrated that each of the five genes is essential for T4MO activity. Other evidence presented indicated that none of the tmo genes was involved in the regulation of the tmo gene cluster, in the control of substrate transport for the T4MO system, or in major processing of the products of the tmo genes. It was tentatively concluded that the tmoABCDE genes encode structural polypeptides of the T4MO enzyme system. One of the tmo genes was tentatively identified as a ferredoxin gene.
Full text
PDF










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bagdasarian M. M., Amann E., Lurz R., Rückert B., Bagdasarian M. Activity of the hybrid trp-lac (tac) promoter of Escherichia coli in Pseudomonas putida. Construction of broad-host-range, controlled-expression vectors. Gene. 1983 Dec;26(2-3):273–282. doi: 10.1016/0378-1119(83)90197-x. [DOI] [PubMed] [Google Scholar]
- Bagdasarian M., Lurz R., Rückert B., Franklin F. C., Bagdasarian M. M., Frey J., Timmis K. N. Specific-purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene. 1981 Dec;16(1-3):237–247. doi: 10.1016/0378-1119(81)90080-9. [DOI] [PubMed] [Google Scholar]
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Chakrabarty A. M., Chou G., Gunsalus I. C. Genetic regulation of octane dissimilation plasmid in Pseudomonas. Proc Natl Acad Sci U S A. 1973 Apr;70(4):1137–1140. doi: 10.1073/pnas.70.4.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline J. F., Hoffman B. M., Mims W. B., LaHaie E., Ballou D. P., Fee J. A. Evidence for N coordination to Fe in the [2Fe-2S] clusters of Thermus Rieske protein and phthalate dioxygenase from Pseudomonas. J Biol Chem. 1985 Mar 25;260(6):3251–3254. [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhaese P., De Greve H., Decraemer H., Schell J., Van Montagu M. Rapid mapping of transposon insertion and deletion mutations in the large Ti-plasmids of Agrobacterium tumefaciens. Nucleic Acids Res. 1979 Dec 11;7(7):1837–1849. doi: 10.1093/nar/7.7.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditta G., Stanfield S., Corbin D., Helinski D. R. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ensley B. D., Ratzkin B. J., Osslund T. D., Simon M. J., Wackett L. P., Gibson D. T. Expression of naphthalene oxidation genes in Escherichia coli results in the biosynthesis of indigo. Science. 1983 Oct 14;222(4620):167–169. doi: 10.1126/science.6353574. [DOI] [PubMed] [Google Scholar]
- Figurski D. H., Helinski D. R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geary P. J., Saboowalla F., Patil D., Cammack R. An investigation of the iron-sulphur proteins of benzene dioxygenase from Pseudomonas putida by electron-spin-resonance spectroscopy. Biochem J. 1984 Feb 1;217(3):667–673. doi: 10.1042/bj2170667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson D. T., Hensley M., Yoshioka H., Mabry T. J. Formation of (+)-cis-2,3-dihydroxy-1-methylcyclohexa-4,6-diene from toluene by Pseudomonas putida. Biochemistry. 1970 Mar 31;9(7):1626–1630. doi: 10.1021/bi00809a023. [DOI] [PubMed] [Google Scholar]
- Hunkapiller M. W., Lujan E., Ostrander F., Hood L. E. Isolation of microgram quantities of proteins from polyacrylamide gels for amino acid sequence analysis. Methods Enzymol. 1983;91:227–236. doi: 10.1016/s0076-6879(83)91019-4. [DOI] [PubMed] [Google Scholar]
- Irie S., Doi S., Yorifuji T., Takagi M., Yano K. Nucleotide sequencing and characterization of the genes encoding benzene oxidation enzymes of Pseudomonas putida. J Bacteriol. 1987 Nov;169(11):5174–5179. doi: 10.1128/jb.169.11.5174-5179.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston J. B., Gunsalus I. C. Isolation of metabolic plasmid DNA from Pseudomonas putida. Biochem Biophys Res Commun. 1977 Mar 7;75(1):13–19. doi: 10.1016/0006-291x(77)91282-7. [DOI] [PubMed] [Google Scholar]
- Klein M. L., Bartley T. D., Davis J. M., Whiteley D. W., Lu H. S. Isolation and structural characterization of three isoforms of recombinant consensus alpha interferon. Arch Biochem Biophys. 1990 Feb 1;276(2):531–537. doi: 10.1016/0003-9861(90)90755-n. [DOI] [PubMed] [Google Scholar]
- Kobal V. M., Gibson D. T., Davis R. E., Garza A. X-ray determination of the absolute stereochemistry of the initial oxidation product formed from toluene by Pseudomonas puida 39-D. J Am Chem Soc. 1973 Jun 27;95(13):4420–4421. doi: 10.1021/ja00794a048. [DOI] [PubMed] [Google Scholar]
- LENNOX E. S. Transduction of linked genetic characters of the host by bacteriophage P1. Virology. 1955 Jul;1(2):190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lu H. S., Lin F. K., Chao L., Chao J. Human urinary kallikrein. Complete amino acid sequence and sites of glycosylation. Int J Pept Protein Res. 1989 Apr;33(4):237–249. doi: 10.1111/j.1399-3011.1989.tb01277.x. [DOI] [PubMed] [Google Scholar]
- Mandel M., Higa A. Calcium-dependent bacteriophage DNA infection. J Mol Biol. 1970 Oct 14;53(1):159–162. doi: 10.1016/0022-2836(70)90051-3. [DOI] [PubMed] [Google Scholar]
- Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem. 1987 Jul 25;262(21):10035–10038. [PubMed] [Google Scholar]
- Morrice N., Geary P., Cammack R., Harris A., Beg F., Aitken A. Primary structure of protein B from Pseudomonas putida, member of a new class of 2Fe-2S ferredoxins. FEBS Lett. 1988 Apr 25;231(2):336–340. doi: 10.1016/0014-5793(88)80845-7. [DOI] [PubMed] [Google Scholar]
- Nordlund I., Powlowski J., Shingler V. Complete nucleotide sequence and polypeptide analysis of multicomponent phenol hydroxylase from Pseudomonas sp. strain CF600. J Bacteriol. 1990 Dec;172(12):6826–6833. doi: 10.1128/jb.172.12.6826-6833.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palleroni N. J., Doudoroff M., Stanier R. Y., Solánes R. E., Mandel M. Taxonomy of the aerobic pseudomonads: the properties of the Pseudomonas stutzeri group. J Gen Microbiol. 1970 Feb;60(2):215–231. doi: 10.1099/00221287-60-2-215. [DOI] [PubMed] [Google Scholar]
- Platt T. Transcription termination and the regulation of gene expression. Annu Rev Biochem. 1986;55:339–372. doi: 10.1146/annurev.bi.55.070186.002011. [DOI] [PubMed] [Google Scholar]
- Powlowski J., Shingler V. In vitro analysis of polypeptide requirements of multicomponent phenol hydroxylase from Pseudomonas sp. strain CF600. J Bacteriol. 1990 Dec;172(12):6834–6840. doi: 10.1128/jb.172.12.6834-6840.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SUSSMAN R., JACOB F. [On a thermosensitive repression system in the Escherichia coli lambda bacteriophage]. C R Hebd Seances Acad Sci. 1962 Feb 19;254:1517–1519. [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields M. S., Montgomery S. O., Chapman P. J., Cuskey S. M., Pritchard P. H. Novel pathway of toluene catabolism in the trichloroethylene-degrading bacterium g4. Appl Environ Microbiol. 1989 Jun;55(6):1624–1629. doi: 10.1128/aem.55.6.1624-1629.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian V., Liu T. N., Yeh W. K., Serdar C. M., Wackett L. P., Gibson D. T. Purification and properties of ferredoxinTOL. A component of toluene dioxygenase from Pseudomonas putida F1. J Biol Chem. 1985 Feb 25;260(4):2355–2363. [PubMed] [Google Scholar]
- Whited G. M., Gibson D. T. Separation and partial characterization of the enzymes of the toluene-4-monooxygenase catabolic pathway in Pseudomonas mendocina KR1. J Bacteriol. 1991 May;173(9):3017–3020. doi: 10.1128/jb.173.9.3017-3020.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whited G. M., Gibson D. T. Toluene-4-monooxygenase, a three-component enzyme system that catalyzes the oxidation of toluene to p-cresol in Pseudomonas mendocina KR1. J Bacteriol. 1991 May;173(9):3010–3016. doi: 10.1128/jb.173.9.3010-3016.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worsey M. J., Williams P. A. Metabolism of toluene and xylenes by Pseudomonas (putida (arvilla) mt-2: evidence for a new function of the TOL plasmid. J Bacteriol. 1975 Oct;124(1):7–13. doi: 10.1128/jb.124.1.7-13.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Yen K. M. Construction of cloning cartridges for development of expression vectors in gram-negative bacteria. J Bacteriol. 1991 Sep;173(17):5328–5335. doi: 10.1128/jb.173.17.5328-5335.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen K. M., Gunsalus I. C. Plasmid gene organization: naphthalene/salicylate oxidation. Proc Natl Acad Sci U S A. 1982 Feb;79(3):874–878. doi: 10.1073/pnas.79.3.874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen K. M., Gunsalus I. C. Regulation of naphthalene catabolic genes of plasmid NAH7. J Bacteriol. 1985 Jun;162(3):1008–1013. doi: 10.1128/jb.162.3.1008-1013.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziffer H., Jerina D. M., Gibson D. T., Kobal V. M. Absolute stereochemistry of the (+)-cis-1,2-dihydroxy-3-methylcyclohexa-3,5-diene produced from toluene by Pseudomonas putida. J Am Chem Soc. 1973 Jun 13;95(12):4048–4049. doi: 10.1021/ja00793a036. [DOI] [PubMed] [Google Scholar]
- Zylstra G. J., Gibson D. T. Toluene degradation by Pseudomonas putida F1. Nucleotide sequence of the todC1C2BADE genes and their expression in Escherichia coli. J Biol Chem. 1989 Sep 5;264(25):14940–14946. [PubMed] [Google Scholar]



