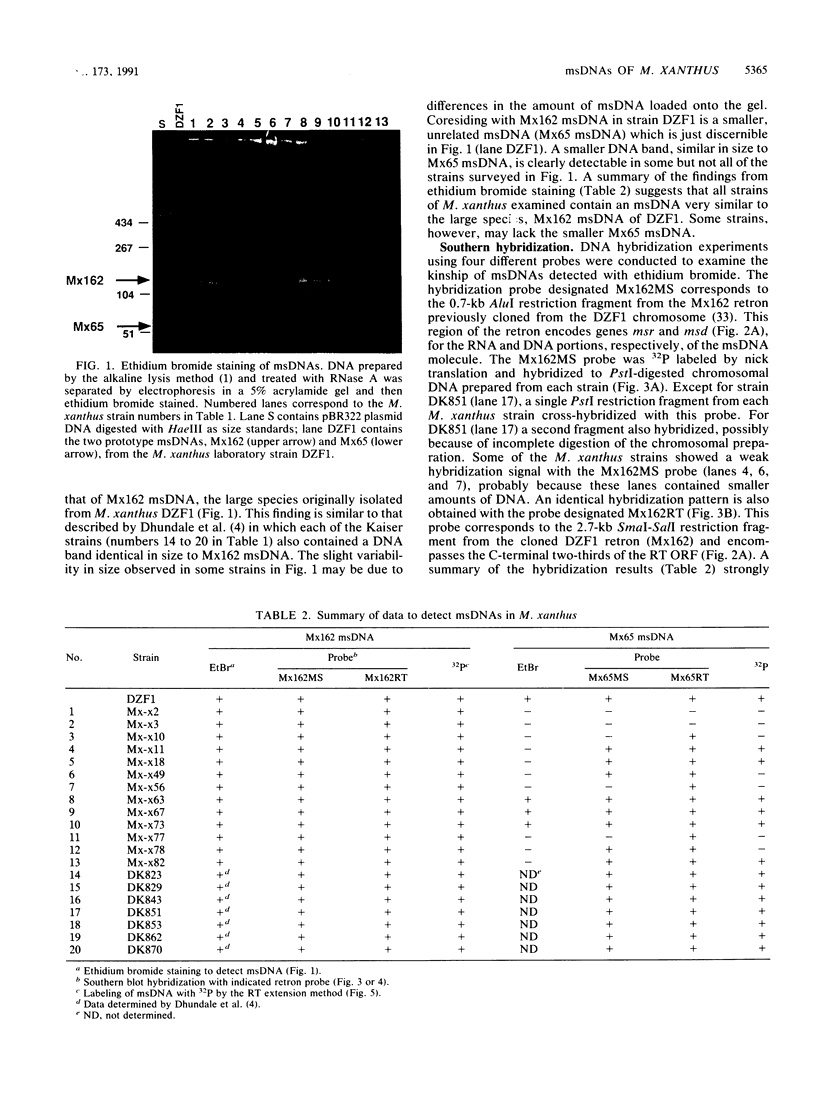
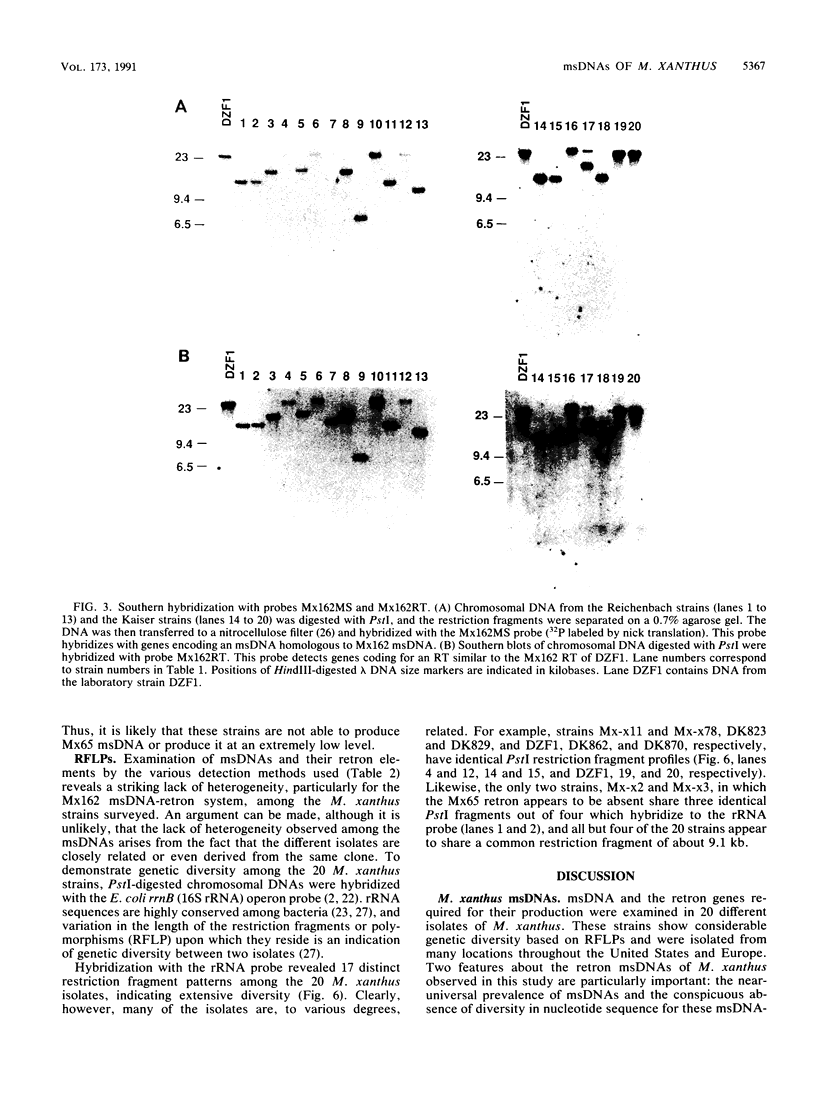
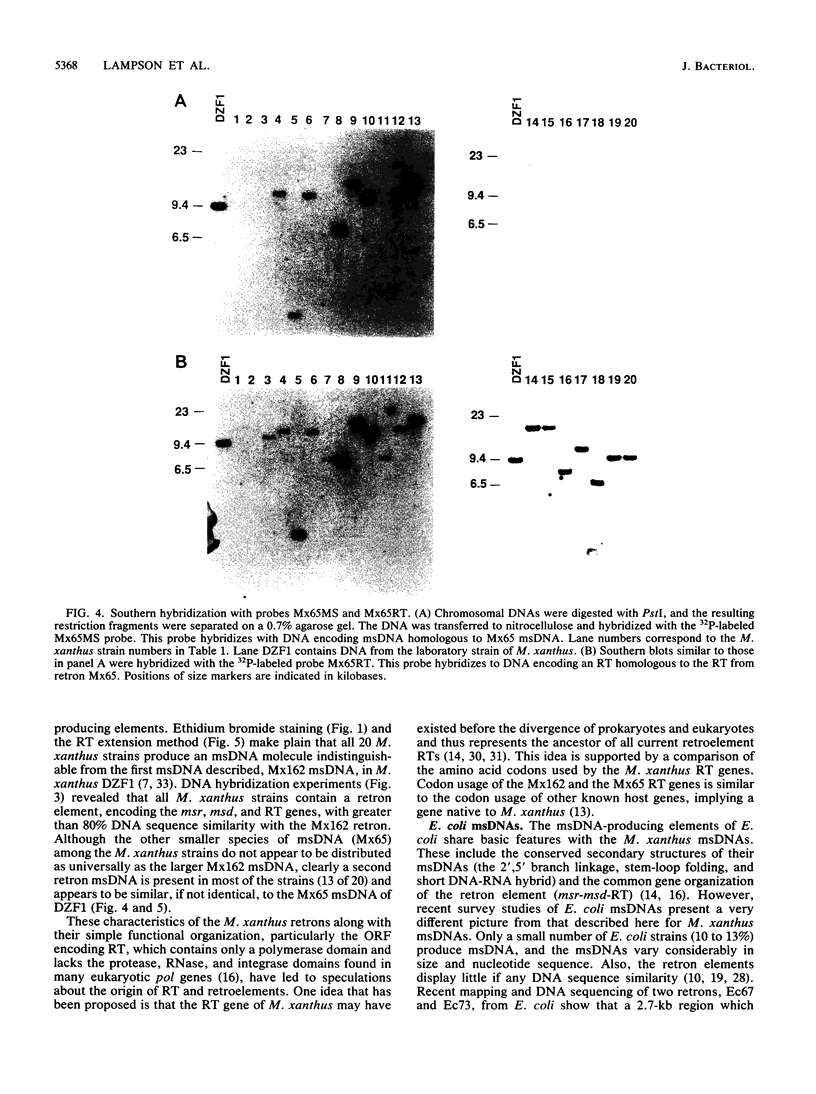
Abstract
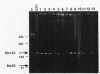
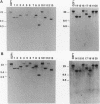
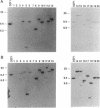
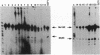
Twenty different isolates of the soil bacterium Myxococcus xanthus were examined for the presence of multicopy single-stranded DNA (msDNA)-producing retroelements, or retrons. Each strain was analyzed by ethidium bromide staining for msDNA, 32P labeling of the msDNA molecule by the reverse transcriptase (RT) extension method, and DNA hybridization experiments with probes derived from two retrons, Mx162 and Mx65, previously cloned from M. xanthus DZF1. These analyses revealed that all M. xanthus strains contain an msDNA very similar to Mx162 msDNA, and 13 strains also contain a second smaller msDNA very similar to Mx65 msDNA. In addition, the strains contained retron-encoded genes msr and msd, which code for msDNA, and a gene for RT responsible for the synthesis of msDNA. These genes show greater than 80% nucleotide sequence similarity to retrons Mx162 or Mx65. The near-ubiquitous occurrence of msDNA retrons among M. xanthus strains and their homogeneous nature are in marked contrast to the highly diverse but rarely occurring msDNA-producing elements of Escherichia coli. The possible origin and evolution of RT and retron elements is discussed in view of these findings.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosius J., Dull T. J., Sleeter D. D., Noller H. F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981 May 15;148(2):107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- Campos J. M., Geisselsoder J., Zusman D. R. Isolation of bacteriophage MX4, a generalized transducing phage for Myxococcus xanthus. J Mol Biol. 1978 Feb 25;119(2):167–178. doi: 10.1016/0022-2836(78)90431-x. [DOI] [PubMed] [Google Scholar]
- DWORKIN M. Nutritional requirements for vegetative growth of Myxococcus xanthus. J Bacteriol. 1962 Aug;84:250–257. doi: 10.1128/jb.84.2.250-257.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhundale A. R., Furuichi T., Inouye S., Inouye M. Distribution of multicopy single-stranded DNA among myxobacteria and related species. J Bacteriol. 1985 Nov;164(2):914–917. doi: 10.1128/jb.164.2.914-917.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhundale A., Furuichi T., Inouye M., Inouye S. Mutations that affect production of branched RNA-linked msDNA in Myxococcus xanthus. J Bacteriol. 1988 Dec;170(12):5620–5624. doi: 10.1128/jb.170.12.5620-5624.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhundale A., Inouye M., Inouye S. A new species of multicopy single-stranded DNA from Myxococcus xanthus with conserved structural features. J Biol Chem. 1988 Jun 25;263(18):9055–9058. [PubMed] [Google Scholar]
- Dhundale A., Lampson B., Furuichi T., Inouye M., Inouye S. Structure of msDNA from Myxococcus xanthus: evidence for a long, self-annealing RNA precursor for the covalently linked, branched RNA. Cell. 1987 Dec 24;51(6):1105–1112. doi: 10.1016/0092-8674(87)90596-4. [DOI] [PubMed] [Google Scholar]
- Furuichi T., Dhundale A., Inouye M., Inouye S. Branched RNA covalently linked to the 5' end of a single-stranded DNA in Stigmatella aurantiaca: structure of msDNA. Cell. 1987 Jan 16;48(1):47–53. doi: 10.1016/0092-8674(87)90354-0. [DOI] [PubMed] [Google Scholar]
- Herzer P. J., Inouye S., Inouye M., Whittam T. S. Phylogenetic distribution of branched RNA-linked multicopy single-stranded DNA among natural isolates of Escherichia coli. J Bacteriol. 1990 Nov;172(11):6175–6181. doi: 10.1128/jb.172.11.6175-6181.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu M. Y., Inouye M., Inouye S. Retron for the 67-base multicopy single-stranded DNA from Escherichia coli: a potential transposable element encoding both reverse transcriptase and Dam methylase functions. Proc Natl Acad Sci U S A. 1990 Dec;87(23):9454–9458. doi: 10.1073/pnas.87.23.9454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye M., Inouye S. Retroelements in bacteria. Trends Biochem Sci. 1991 Jan;16(1):18–21. doi: 10.1016/0968-0004(91)90010-s. [DOI] [PubMed] [Google Scholar]
- Inouye S., Herzer P. J., Inouye M. Two independent retrons with highly diverse reverse transcriptases in Myxococcus xanthus. Proc Natl Acad Sci U S A. 1990 Feb;87(3):942–945. doi: 10.1073/pnas.87.3.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye S., Hsu M. Y., Eagle S., Inouye M. Reverse transcriptase associated with the biosynthesis of the branched RNA-linked msDNA in Myxococcus xanthus. Cell. 1989 Feb 24;56(4):709–717. doi: 10.1016/0092-8674(89)90593-x. [DOI] [PubMed] [Google Scholar]
- Lampson B. C., Inouye M., Inouye S. Reverse transcriptase with concomitant ribonuclease H activity in the cell-free synthesis of branched RNA-linked msDNA of Myxococcus xanthus. Cell. 1989 Feb 24;56(4):701–707. doi: 10.1016/0092-8674(89)90592-8. [DOI] [PubMed] [Google Scholar]
- Lampson B. C., Inouye S., Inouye M. msDNA of bacteria. Prog Nucleic Acid Res Mol Biol. 1991;40:1–24. doi: 10.1016/s0079-6603(08)60838-7. [DOI] [PubMed] [Google Scholar]
- Lampson B. C., Sun J., Hsu M. Y., Vallejo-Ramirez J., Inouye S., Inouye M. Reverse transcriptase in a clinical strain of Escherichia coli: production of branched RNA-linked msDNA. Science. 1989 Feb 24;243(4894 Pt 1):1033–1038. doi: 10.1126/science.2466332. [DOI] [PubMed] [Google Scholar]
- Lampson B. C., Viswanathan M., Inouye M., Inouye S. Reverse transcriptase from Escherichia coli exists as a complex with msDNA and is able to synthesize double-stranded DNA. J Biol Chem. 1990 May 25;265(15):8490–8496. [PubMed] [Google Scholar]
- Lim D., Gomes T. A., Maas W. K. Distribution of msDNAs among serotypes of enteropathogenic Escherichia coli strains. Mol Microbiol. 1990 Oct;4(10):1711–1714. doi: 10.1111/j.1365-2958.1990.tb00548.x. [DOI] [PubMed] [Google Scholar]
- Lim D., Maas W. K. Mapping of the msDNA operon in the chromosome of Escherichia coli B. Mol Microbiol. 1990 Dec;4(12):2201–2204. doi: 10.1111/j.1365-2958.1990.tb00582.x. [DOI] [PubMed] [Google Scholar]
- Lim D., Maas W. K. Reverse transcriptase-dependent synthesis of a covalently linked, branched DNA-RNA compound in E. coli B. Cell. 1989 Mar 10;56(5):891–904. doi: 10.1016/0092-8674(89)90693-4. [DOI] [PubMed] [Google Scholar]
- Lindahl L., Zengel J. M. Ribosomal genes in Escherichia coli. Annu Rev Genet. 1986;20:297–326. doi: 10.1146/annurev.ge.20.120186.001501. [DOI] [PubMed] [Google Scholar]
- Moore R. L., McCarthy B. J. Comparative study of ribosomal ribonucleic acid cistrons in enterobacteria and myxobacteria. J Bacteriol. 1967 Oct;94(4):1066–1074. doi: 10.1128/jb.94.4.1066-1074.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H., Selander R. K. Standard reference strains of Escherichia coli from natural populations. J Bacteriol. 1984 Feb;157(2):690–693. doi: 10.1128/jb.157.2.690-693.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stull T. L., LiPuma J. J., Edlind T. D. A broad-spectrum probe for molecular epidemiology of bacteria: ribosomal RNA. J Infect Dis. 1988 Feb;157(2):280–286. doi: 10.1093/infdis/157.2.280. [DOI] [PubMed] [Google Scholar]
- Sun J., Herzer P. J., Weinstein M. P., Lampson B. C., Inouye M., Inouye S. Extensive diversity of branched-RNA-linked multicopy single-stranded DNAs in clinical strains of Escherichia coli. Proc Natl Acad Sci U S A. 1989 Sep;86(18):7208–7212. doi: 10.1073/pnas.86.18.7208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Inouye M., Inouye S. Association of a retroelement with a P4-like cryptic prophage (retronphage phi R73) integrated into the selenocystyl tRNA gene of Escherichia coli. J Bacteriol. 1991 Jul;173(13):4171–4181. doi: 10.1128/jb.173.13.4171-4181.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temin H. M. Reverse transcriptases. Retrons in bacteria. Nature. 1989 May 25;339(6222):254–255. doi: 10.1038/339254a0. [DOI] [PubMed] [Google Scholar]
- Varmus H. E. Reverse transcription in bacteria. Cell. 1989 Mar 10;56(5):721–724. doi: 10.1016/0092-8674(89)90673-9. [DOI] [PubMed] [Google Scholar]
- Xiong Y., Eickbush T. H. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 1990 Oct;9(10):3353–3362. doi: 10.1002/j.1460-2075.1990.tb07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee T., Furuichi T., Inouye S., Inouye M. Multicopy single-stranded DNA isolated from a gram-negative bacterium, Myxococcus xanthus. Cell. 1984 Aug;38(1):203–209. doi: 10.1016/0092-8674(84)90541-5. [DOI] [PubMed] [Google Scholar]