Abstract
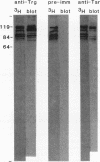
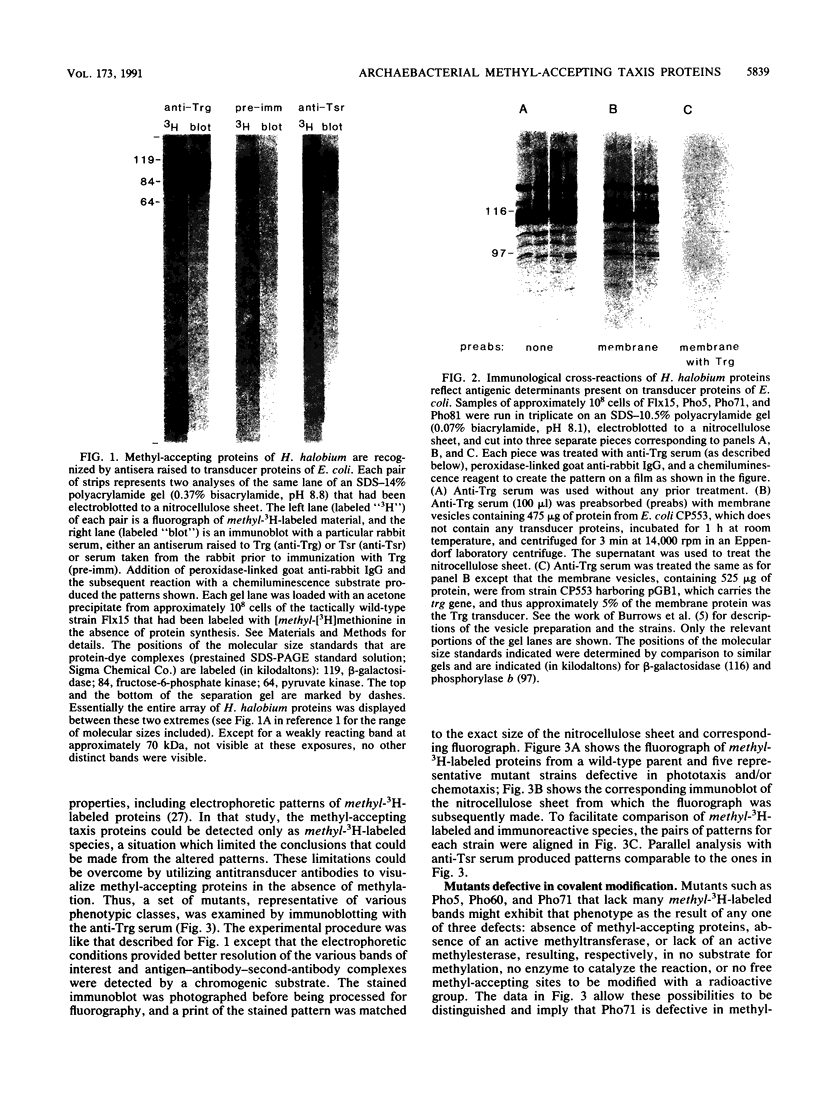
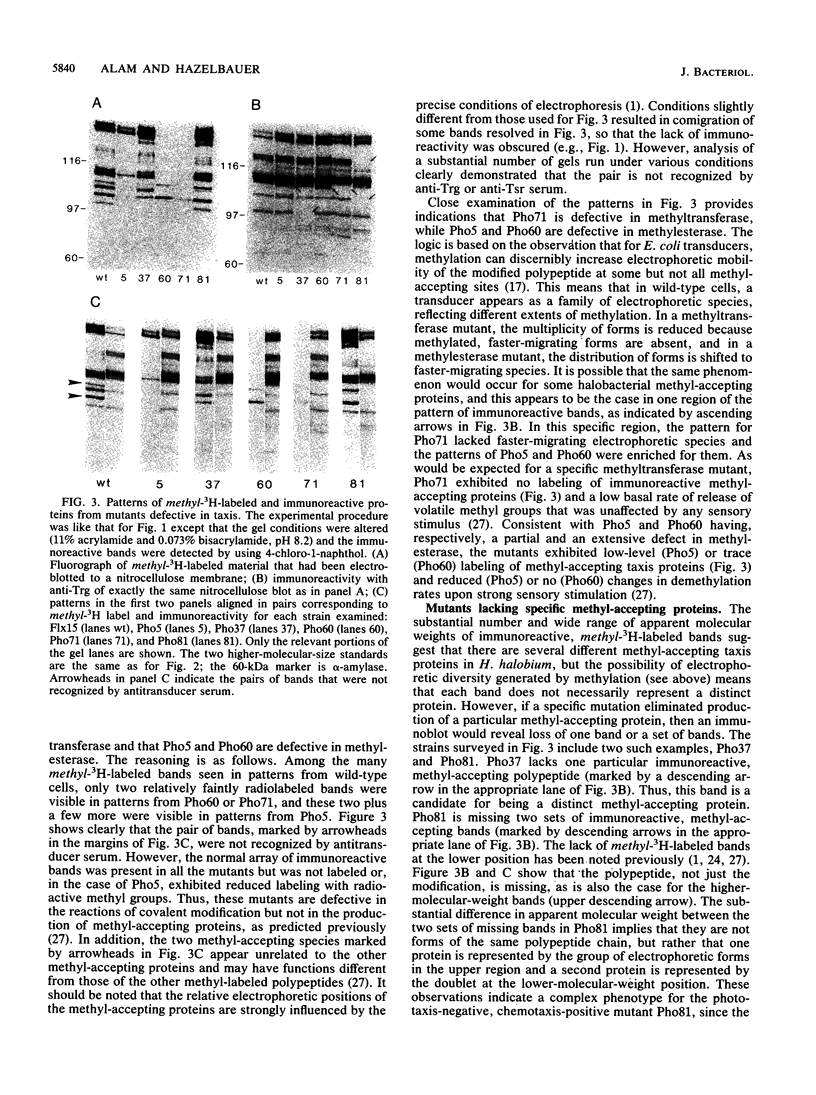
A number of eubacterial species contain methyl-accepting taxis proteins that are antigenically and thus structurally related to the well-characterized methyl-accepting chemotaxis proteins of Escherichia coli. Recent studies of the archaebacterium Halobacterium halobium have characterized methyl-accepting taxis proteins that in some ways resemble and in other ways differ from the analogous eubacterial proteins. We used immunoblotting with antisera raised to E. coli transducers to probe shared structural features of methyl-accepting proteins from archaebacteria and eubacteria and found substantial antigenic relationships. This implies that the genes for the contemporary methyl-accepting proteins are related through an ancestral gene that existed before the divergence of arachaebacteria and eubacteria. Analysis by immunoblot of mutants of H. halobium defective in taxis revealed that some strains were deficient in covalent modification of methyl-accepting proteins although the proteins themselves were present, while other strains appeared to be missing specific methyl-accepting proteins.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alam M., Lebert M., Oesterhelt D., Hazelbauer G. L. Methyl-accepting taxis proteins in Halobacterium halobium. EMBO J. 1989 Feb;8(2):631–639. doi: 10.1002/j.1460-2075.1989.tb03418.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollinger J., Park C., Harayama S., Hazelbauer G. L. Structure of the Trg protein: Homologies with and differences from other sensory transducers of Escherichia coli. Proc Natl Acad Sci U S A. 1984 Jun;81(11):3287–3291. doi: 10.1073/pnas.81.11.3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borkovich K. A., Kaplan N., Hess J. F., Simon M. I. Transmembrane signal transduction in bacterial chemotaxis involves ligand-dependent activation of phosphate group transfer. Proc Natl Acad Sci U S A. 1989 Feb;86(4):1208–1212. doi: 10.1073/pnas.86.4.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd A., Kendall K., Simon M. I. Structure of the serine chemoreceptor in Escherichia coli. Nature. 1983 Feb 17;301(5901):623–626. doi: 10.1038/301623a0. [DOI] [PubMed] [Google Scholar]
- Burrows G. G., Newcomer M. E., Hazelbauer G. L. Purification of receptor protein Trg by exploiting a property common to chemotactic transducers of Escherichia coli. J Biol Chem. 1989 Oct 15;264(29):17309–17315. [PubMed] [Google Scholar]
- Craven R. C., Montie T. C. Chemotaxis of Pseudomonas aeruginosa: involvement of methylation. J Bacteriol. 1983 May;154(2):780–786. doi: 10.1128/jb.154.2.780-786.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl M. K., Boos W., Manson M. D. Evolution of chemotactic-signal transducers in enteric bacteria. J Bacteriol. 1989 May;171(5):2361–2371. doi: 10.1128/jb.171.5.2361-2371.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engström P., Nowlin D., Bollinger J., Magnuson N., Hazelbauer G. L. Limited homology between trg and the other transducer proteins of Escherichia coli. J Bacteriol. 1983 Dec;156(3):1268–1274. doi: 10.1128/jb.156.3.1268-1274.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood C. S. A methyl-accepting protein is involved in benzoate taxis in Pseudomonas putida. J Bacteriol. 1989 Sep;171(9):4603–4608. doi: 10.1128/jb.171.9.4603-4608.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazelbauer G. L. The bacterial chemosensory system. Can J Microbiol. 1988 Apr;34(4):466–474. doi: 10.1139/m88-080. [DOI] [PubMed] [Google Scholar]
- Hirota N. Methylation and demethylation of solubilized chemoreceptors from thermophilic bacterium PS-3. J Biochem. 1986 Feb;99(2):349–356. doi: 10.1093/oxfordjournals.jbchem.a135489. [DOI] [PubMed] [Google Scholar]
- Kathariou S., Greenberg E. P. Chemoattractants elicit methylation of specific polypeptides in Spirochaeta aurantia. J Bacteriol. 1983 Oct;156(1):95–100. doi: 10.1128/jb.156.1.95-100.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krikos A., Mutoh N., Boyd A., Simon M. I. Sensory transducers of E. coli are composed of discrete structural and functional domains. Cell. 1983 Jun;33(2):615–622. doi: 10.1016/0092-8674(83)90442-7. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Nowlin D. M., Bollinger J., Hazelbauer G. L. Site-directed mutations altering methyl-accepting residues of a sensory transducer protein. Proteins. 1988;3(2):102–112. doi: 10.1002/prot.340030205. [DOI] [PubMed] [Google Scholar]
- Oesterhelt D., Stoeckenius W. Isolation of the cell membrane of Halobacterium halobium and its fractionation into red and purple membrane. Methods Enzymol. 1974;31:667–678. doi: 10.1016/0076-6879(74)31072-5. [DOI] [PubMed] [Google Scholar]
- Randall L. L., Hardy S. J. Synthesis of exported proteins by membrane-bound polysomes from Escherichia coli. Eur J Biochem. 1977 May 2;75(1):43–53. doi: 10.1111/j.1432-1033.1977.tb11502.x. [DOI] [PubMed] [Google Scholar]
- Russo A. F., Koshland D. E., Jr Separation of signal transduction and adaptation functions of the aspartate receptor in bacterial sensing. Science. 1983 Jun 3;220(4601):1016–1020. doi: 10.1126/science.6302843. [DOI] [PubMed] [Google Scholar]
- Shaw P., Gomes S. L., Sweeney K., Ely B., Shapiro L. Methylation involved in chemotaxis is regulated during Caulobacter differentiation. Proc Natl Acad Sci U S A. 1983 Sep;80(17):5261–5265. [PMC free article] [PubMed] [Google Scholar]
- Sockett R. E., Armitage J. P., Evans M. C. Methylation-independent and methylation-dependent chemotaxis in Rhodobacter sphaeroides and Rhodospirillum rubrum. J Bacteriol. 1987 Dec;169(12):5808–5814. doi: 10.1128/jb.169.12.5808-5814.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich E. N., Hasselbacher C. A., Spudich J. L. Methyl-accepting protein associated with bacterial sensory rhodopsin I. J Bacteriol. 1988 Sep;170(9):4280–4285. doi: 10.1128/jb.170.9.4280-4285.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich E. N., Spudich J. L. Control of transmembrane ion fluxes to select halorhodopsin-deficient and other energy-transduction mutants of Halobacterium halobium. Proc Natl Acad Sci U S A. 1982 Jul;79(14):4308–4312. doi: 10.1073/pnas.79.14.4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich E. N., Takahashi T., Spudich J. L. Sensory rhodopsins I and II modulate a methylation/demethylation system in Halobacterium halobium phototaxis. Proc Natl Acad Sci U S A. 1989 Oct;86(20):7746–7750. doi: 10.1073/pnas.86.20.7746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundberg S. A., Alam M., Lebert M., Spudich J. L., Oesterhelt D., Hazelbauer G. L. Characterization of Halobacterium halobium mutants defective in taxis. J Bacteriol. 1990 May;172(5):2328–2335. doi: 10.1128/jb.172.5.2328-2335.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundberg S. A., Bogomolni R. A., Spudich J. L. Selection and properties of phototaxis-deficient mutants of Halobacterium halobium. J Bacteriol. 1985 Oct;164(1):282–287. doi: 10.1128/jb.164.1.282-287.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah A. H., Ordal G. W. In vivo and in vitro chemotactic methylation in Bacillus subtilis. J Bacteriol. 1981 Feb;145(2):958–965. doi: 10.1128/jb.145.2.958-965.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R. Bacterial evolution. Microbiol Rev. 1987 Jun;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]