Abstract
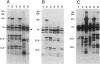
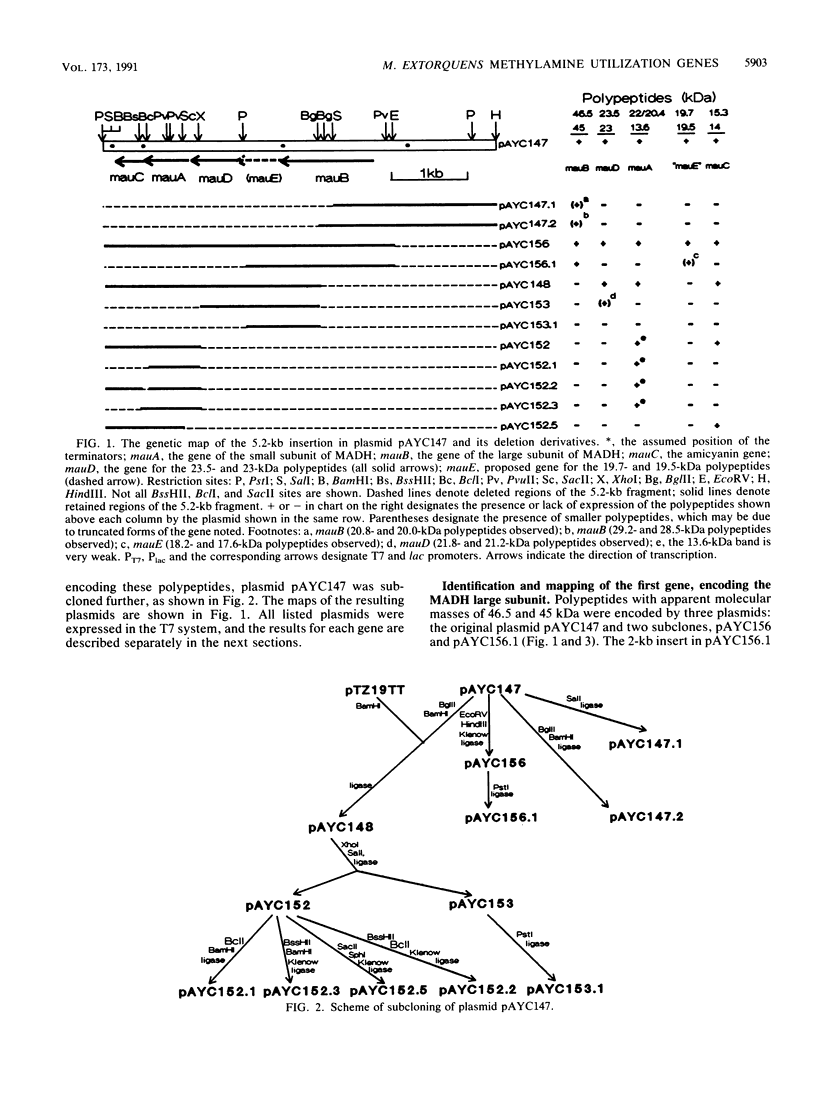
An isolated 5.2-kb fragment of Methylobacterium extorquens AM1 DNA was found to contain a gene cluster involved in methylamine utilization. Analysis of polypeptides synthesized in an Escherichia coli T7 expression system showed that five genes were present. Two of the genes encoded the large and small subunits of methylamine dehydrogenase, and a third encoded amicyanin, the presumed electron acceptor for methylamine dehydrogenase, but the function of the other two genes is not known. The order on the 5.2-kb fragment was found to be large-subunit gene, the two genes of unknown function, small-subunit gene, amicyanin gene. The gene for azurin, another possible electron acceptor in methylamine oxidation, does not appear to be present within this cluster of methylamine utilization genes.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ambler R. P., Tobari J. The primary structures of Pseudomonas AM1 amicyanin and pseudoazurin. Two new sequence classes of blue copper proteins. Biochem J. 1985 Dec 1;232(2):451–457. doi: 10.1042/bj2320451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson D. J., Lidstrom M. E. The moxFG region encodes four polypeptides in the methanol-oxidizing bacterium Methylobacterium sp. strain AM1. J Bacteriol. 1988 May;170(5):2254–2262. doi: 10.1128/jb.170.5.2254-2262.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson D. J., Morris C. J., Nunn D. N., Anthony C., Lidstrom M. E. Nucleotide sequence of the Methylobacterium extorquens AM1 moxF and moxJ genes involved in methanol oxidation. Gene. 1990 May 31;90(1):173–176. doi: 10.1016/0378-1119(90)90457-3. [DOI] [PubMed] [Google Scholar]
- Bellion E., Hersh L. B. Methylamine metabolism in a pseudomonas species. Arch Biochem Biophys. 1972 Nov;153(1):368–374. doi: 10.1016/0003-9861(72)90457-2. [DOI] [PubMed] [Google Scholar]
- Bellion E., Kim Y. S. Catabolite repression of isocitrate lyase in methylamine-grown Pseudomonas MA. Effect of carbon and nitrogen sources. Biochim Biophys Acta. 1978 Jul 17;541(4):425–434. doi: 10.1016/0304-4165(78)90152-6. [DOI] [PubMed] [Google Scholar]
- Chandrasekar R., Klapper M. H. Methylamine dehydrogenase and cytochrome c552 from the bacterium W3A1. J Biol Chem. 1986 Mar 15;261(8):3616–3619. [PubMed] [Google Scholar]
- Chistoserdov A. Y., Lidstrom M. E. The small-subunit polypeptide of methylamine dehydrogenase from Methylobacterium extorquens AM1 has an unusual leader sequence. J Bacteriol. 1991 Sep;173(18):5909–5913. doi: 10.1128/jb.173.18.5909-5913.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chistoserdov A. Y., Tsygankov Y. D., Lidstrom M. E. Cloning and sequencing of the structural gene for the small subunit of methylamine dehydrogenase from Methylobacterium extorquens AM1: evidence for two tryptophan residues involved in the active center. Biochem Biophys Res Commun. 1990 Oct 15;172(1):211–216. doi: 10.1016/s0006-291x(05)80195-0. [DOI] [PubMed] [Google Scholar]
- Day D. J., Nunn D. N., Anthony C. Characterization of a novel soluble c-type cytochrome in a moxD mutant of Methylobacterium extorquens AM1. J Gen Microbiol. 1990 Jan;136(1):181–188. doi: 10.1099/00221287-136-1-181. [DOI] [PubMed] [Google Scholar]
- Eady R. R., Large P. J. Microbial oxidation of amines. Spectral and kinetic properties of the primary amine dehydrogenase of Pseudomonas AM1. Biochem J. 1971 Aug;123(5):757–771. doi: 10.1042/bj1230757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eady R. R., Large P. J. Purification and properties of an amine dehydrogenase from Pseudomonas AM1 and its role in growth on methylamine. Biochem J. 1968 Jan;106(1):245–255. doi: 10.1042/bj1060245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukumori Y., Yamanaka T. The methylamine oxidizing system of Pseudomonas AM1 reconstituted with purified components. J Biochem. 1987 Feb;101(2):441–445. doi: 10.1093/oxfordjournals.jbchem.a121929. [DOI] [PubMed] [Google Scholar]
- Fulton G. L., Nunn D. N., Lidstrom M. E. Molecular cloning of a malyl coenzyme A lyase gene from Pseudomonas sp. strain AM1, a facultative methylotroph. J Bacteriol. 1984 Nov;160(2):718–723. doi: 10.1128/jb.160.2.718-723.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hersh L. B., Peterson J. A., Thompson A. A. An N-methyl glutamate dehydrogenase from Pseudomonas M.A. Arch Biochem Biophys. 1971 Jul;145(1):115–120. doi: 10.1016/0003-9861(71)90016-6. [DOI] [PubMed] [Google Scholar]
- Husain M., Davidson V. L. An inducible periplasmic blue copper protein from Paracoccus denitrificans. Purification, properties, and physiological role. J Biol Chem. 1985 Nov 25;260(27):14626–14629. [PubMed] [Google Scholar]
- Husain M., Davidson V. L. Characterization of two inducible periplasmic c-type cytochromes from Paracoccus denitrificans. J Biol Chem. 1986 Jul 5;261(19):8577–8580. [PubMed] [Google Scholar]
- Husain M., Davidson V. L. Purification and properties of methylamine dehydrogenase from Paracoccus denitrificans. J Bacteriol. 1987 Apr;169(4):1712–1717. doi: 10.1128/jb.169.4.1712-1717.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii Y., Hase T., Fukumori Y., Matsubara H., Tobari J. Amino acid sequence studies of the light subunit of methylamine dehydrogenase from Pseudomonas AM1: existence of two residues binding the prosthetic group. J Biochem. 1983 Jan;93(1):107–119. doi: 10.1093/oxfordjournals.jbchem.a134144. [DOI] [PubMed] [Google Scholar]
- Kenney W. C., McIntire W. Characterization of methylamine dehydrogenase from bacterium W3A1. Interaction with reductants and amino-containing compounds. Biochemistry. 1983 Aug 2;22(16):3858–3868. doi: 10.1021/bi00285a022. [DOI] [PubMed] [Google Scholar]
- Kung H. F., Wagner C. Gamma-glutamylmethylamide. A new intermediate in the metabolism of methylamine. J Biol Chem. 1969 Aug 10;244(15):4136–4140. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lawton S. A., Anthony C. The role of blue copper proteins in the oxidation of methylamine by an obligate methylotroph. Biochem J. 1985 Jun 15;228(3):719–726. doi: 10.1042/bj2280719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machlin S. M., Tam P. E., Bastien C. A., Hanson R. S. Genetic and physical analyses of Methylobacterium organophilum XX genes encoding methanol oxidation. J Bacteriol. 1988 Jan;170(1):141–148. doi: 10.1128/jb.170.1.141-148.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinkus K., Kennelly P. J., Rea T., Timkovich R. Purification and properties of Paracoccus denitrificans Azurin. Arch Biochem Biophys. 1980 Feb;199(2):465–472. doi: 10.1016/0003-9861(80)90303-3. [DOI] [PubMed] [Google Scholar]
- Matsumoto T., Hiraoka B. Y., Tobari J. Methylamine dehydrogenase of Pseudomonase sp. J Isolation and properties of the subunits. Biochim Biophys Acta. 1978 Feb 10;522(2):303–310. doi: 10.1016/0005-2744(78)90064-5. [DOI] [PubMed] [Google Scholar]
- Matsumoto T. Methylamine dehydrogenase of Pseudomonas sp. J. Purification and properties. Biochim Biophys Acta. 1978 Feb 10;522(2):291–302. doi: 10.1016/0005-2744(78)90063-3. [DOI] [PubMed] [Google Scholar]
- McIntire W. S., Wemmer D. E., Chistoserdov A., Lidstrom M. E. A new cofactor in a prokaryotic enzyme: tryptophan tryptophylquinone as the redox prosthetic group in methylamine dehydrogenase. Science. 1991 May 10;252(5007):817–824. doi: 10.1126/science.2028257. [DOI] [PubMed] [Google Scholar]
- Mehta R. J. Methylamine dehydrogenase from the obligate methylotroph Methylomonas methylovora. Can J Microbiol. 1977 Apr;23(4):402–406. doi: 10.1139/m77-059. [DOI] [PubMed] [Google Scholar]
- Meinkoth J., Wahl G. Hybridization of nucleic acids immobilized on solid supports. Anal Biochem. 1984 May 1;138(2):267–284. doi: 10.1016/0003-2697(84)90808-x. [DOI] [PubMed] [Google Scholar]
- Nunn D. N., Lidstrom M. E. Isolation and complementation analysis of 10 methanol oxidation mutant classes and identification of the methanol dehydrogenase structural gene of Methylobacterium sp. strain AM1. J Bacteriol. 1986 May;166(2):581–590. doi: 10.1128/jb.166.2.581-590.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PEEL D., QUAYLE J. R. Microbial growth on C1 compounds. I. Isolation and characterization of Pseudomonas AM 1. Biochem J. 1961 Dec;81:465–469. doi: 10.1042/bj0810465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirai S., Matsumoto T., Tobari J. Methylamine dehydrogenase of Pseudomonas AM1. A subunit enzyme. J Biochem. 1978 Jun;83(6):1599–1607. doi: 10.1093/oxfordjournals.jbchem.a132071. [DOI] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Tobari J., Harada Y. Amicyanin: an electron acceptor of methylamine dehydrogenase. Biochem Biophys Res Commun. 1981 Jul 30;101(2):502–508. doi: 10.1016/0006-291x(81)91288-2. [DOI] [PubMed] [Google Scholar]
- Trotsenko Iu A., Loginova N. V., Shishkina V. N. Metabolizm metilamina u gifomikrobov. Dokl Akad Nauk SSSR. 1974 Jul 21;216(6):1413–1415. [PubMed] [Google Scholar]
- van Houwelingen T., Canters G. W., Stobbelaar G., Duine J. A., Frank J., Jr, Tsugita A. Isolation and characterization of a blue copper protein from Thiobacillus versutus. Eur J Biochem. 1985 Nov 15;153(1):75–80. doi: 10.1111/j.1432-1033.1985.tb09268.x. [DOI] [PubMed] [Google Scholar]
- van Spanning R. J., Wansell C. W., Reijnders W. N., Oltmann L. F., Stouthamer A. H. Mutagenesis of the gene encoding amicyanin of Paracoccus denitrificans and the resultant effect on methylamine oxidation. FEBS Lett. 1990 Nov 26;275(1-2):217–220. doi: 10.1016/0014-5793(90)81475-4. [DOI] [PubMed] [Google Scholar]