Summary
The genetic basis of myotonic dystrophy type 1 (DM1) is a CTG expansion in the 3' untranslated region (UTR) of DMPK. The pathogenic mechanism involves an RNA gain of function in which the repeat containing transcripts accumulate in nuclei and alter the functions of RNA binding proteins such as CUG binding protein 1 (CUGBP1). CUGBP1 levels are increased in DM1 myoblasts, heart and skeletal muscle tissues and in some DM1 mouse models. However, the molecular mechanisms for increased CUGBP1 in DM1 are unclear. Here we demonstrate that expression of DMPK-CUG repeat RNA results in hyper-phosphorylation and stabilization of CUGBP1. CUGBP1 is hyper-phosphorylated in DM1 tissues, cells, and a DM1 mouse model. Activation of PKC is required for CUGBP1 hyper-phosphorylation in DM1 cells and PKCα and βII directly phosphorylate CUGBP1 in vitro. These results indicate that inappropriate activation of the PKC pathway contributes to the pathogenic effects of a non-coding RNA.
Introduction
Myotonic dystrophy (DM) is the most common adult onset muscular dystrophy affecting mainly skeletal muscle, heart, and the CNS (Harper, 2001). Mutations in two genes cause DM. DM type I (DM1) is caused by an expansion of a CTG repeat within the 3' UTR of the DMPK gene and DM2 is caused by a CCTG repeat expansion with intron 1 of the ZNF9 gene (Ranum and Cooper, 2006). The pathogenic mechanism involves a novel RNA gain of function in which repeat containing transcripts from the expanded allele accumulate in nuclei and alter the functions of RNA binding proteins involved in regulating alternative splicing and mRNA translation (Kuyumcu-Martinez and Cooper, 2006). A particularly characteristic feature of DM1 is misregulation of a subset of developmentally regulated alternative splicing events in which the embryonic pattern of splicing occurs in DM1 adult tissues (Ranum and Cooper, 2006). For the muscle specific chloride channel and insulin receptor, expression of the embryonic splicing patterns result in myotonia and insulin resistance, respectively, two key features of the disease (Charlet-B. et al., 2002; Lueck et al., 2007; Mankodi et al., 2002; Savkur et al., 2001).
Expanded CUG repeats mediate their effects on alternative splicing regulation through at least two RNA binding proteins: muscleblind like 1 (MBNL1) and CUGBP1. These proteins are antagonist regulators of splicing events that are misregulated in DM1 such that the splicing patterns observed are consistent with a loss of MBNL1 and a gain of CUGBP1 function (Ho et al., 2004; Lin et al., 2006; Philips et al., 1998; Savkur et al., 2001). Loss of MBNL1 is due to sequestration of the protein on nuclear RNA foci. MBNL1 binds to expanded CUG repeats and colocalizes with RNA foci (Cardani et al., 2006; Fardaei et al., 2001; Fardaei et al., 2002). Consistent with a role for MBNL1 loss of function in DM1 pathogenesis, a knockout of MBNL1 isoforms that can bind to expanded CUG repeats (MBNLΔE3/ΔE3) exhibit many characteristics of DM1 including splicing changes, cataracts, and myotonia (Kanadia et al., 2003; Lin et al., 2006).
Consistent with a gain of CUGBP1 activity in DM1, CUGBP1 steady state levels are increased in DM1 myoblasts, skeletal muscle and heart tissues (Dansithong et al., 2005; Savkur et al., 2001; Timchenko et al., 2001). Results from several studies support the pathogenic role of increased CUGBP1 in DM1 (Dansithong et al., 2005; Ho et al., 2005a; Mahadevan et al., 2006; Savkur et al., 2001; Timchenko et al., 2001; Timchenko et al., 2004). CUGBP1 regulates alternative splicing and translation and pathogenesis is proposed to result from disruption of both functions in DM1 (Ranum and Cooper, 2006). Two different mouse models for CUGBP1 over expression in skeletal muscle exhibited embryonic lethality and myopathy resembling the most severe, congenital form of DM1 (Ho et al., 2005a; Timchenko et al., 2004). One model exhibited mis-splicing events that are observed in individuals with DM1 (Ho et al., 2005a) and the other exhibited altered expression of p21 and MEF2A consistent with altered translational activity (Timchenko et al., 2004). In addition, two recent DM1 mouse models which inducibly express RNA containing CUG repeats in the context of the DMPK 3' UTR exhibited several major DM1 features including increased CUGBP1 expression (Mahadevan et al., 2006; Wang et al., 2007). In one of these, the DMPK-CUG repeat transgene contained only five repeats (Mahadevan et al., 2006). Since five repeats provide a suboptimal binding site for MBNL1, the effects of this toxic RNA were proposed to be due to the elevated CUGBP1 rather than sequestered MBNL1 (Mahadevan et al., 2006).
Unlike MBNL1, CUGBP1 does not colocalize with CUG repeat RNA foci (Wang et al., 2007; Mankodi et al., 2005; Jiang et al., 2004) . CUGBP1 interacts with short single-stranded CUG repeats but not the imperfect double stranded structure of expanded CUG repeats (Timchenko et al., 1996). Therefore, while the mechanism by which MBNL1 is depleted from nuclear pools through sequestration appears straightforward, the mechanism by which CUGBP1 levels increase has been unclear.
In this study we demonstrate that increased CUGBP1 steady state levels in DM1 is due to protein hyper-phosphorylation. We show that expression of DMPK-CUG repeat RNA induces CUGBP1 hyper-phosphorylation, activates protein C kinase (PKC) isoforms and that activation of PKC is required for DMPK-CUG repeat RNA-induced CUGBP1 hyper-phosphorylation and increased steady state levels. Furthermore, CUGBP1 is directly phosphorylated by PKC isozymes in vitro. These results support the hypothesis that expression of the non-coding DMPK-CUG repeat RNA disrupt normal signaling pathways leading to reversion to an embryonic pattern (Ranum and Cooper, 2006).
RESULTS
Expression of expanded CUG repeat mRNA induces hyper-phosphorylation of nuclear CUGBP1
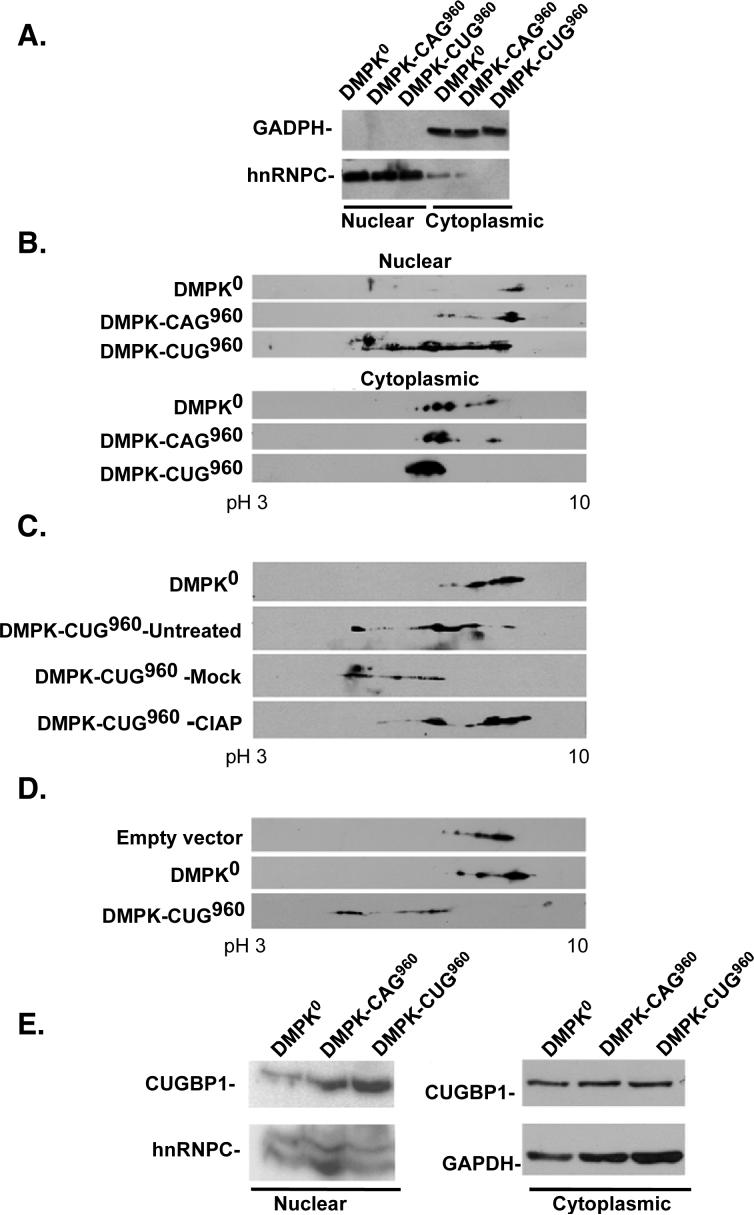
We have previously shown that expression of non coding RNAs containing the DMPK 3' UTR with 960 CUG repeats (DMPK-CUG960 mRNA) induced the DM1 splicing pattern of cTNT and IR minigenes while an identical DMPK mRNA lacking repeats (DMPK0) or containing 960 CAG repeats (DMPK-CAG960) had little effect on cTNT or IR splicing (Ho et al., 2004; Ho et al., 2005b; Philips et al., 1998; Savkur et al., 2001). Mutant cTNT and IR minigene pre-mRNAs that were no longer bound and regulated by CUGBP1 did not respond to DMPK-CUG960 mRNA expression demonstrating a role for CUGBP1 in the trans-dominant effect of DMPK-CUG960 RNA (Philips et al., 1998; Savkur et al., 2001). We used two-dimensional (2D) gel electrophoresis to examine whether expression of expanded CUG repeats induced post-translational modifications of CUGBP1. Following transient expression of DMPK0, DMPK-CUG960, and DMPK-CAG960 mRNAs in COS M6 cells, nuclear and cytoplasmic fractions were analyzed by 2D gel separation followed by western blot for CUGBP1 using the 3B1 monoclonal antibody. In parallel plates, DMPK-CUG960 but neither DMPK-CAG960 nor DMPK0 co-expressed with the cTNT minigene induced cTNT splicing changes as described previously (data not shown). Clean separation of nuclear and cytoplasmic fractions was confirmed by western blots using nuclear (hnRNPC) and cytoplasmic (GAPDH) markers (Figure 1A). The isoelectric point (pI) of endogenous CUGBP1 exhibited a striking acidic shift in nuclear fractions of cells expressing DMPK-CUG960 mRNA compared to cells expressing DMPK-CAG960 or DMPK0 (Figure 1B). There was little change in the pI of CUGBP1 in the cytoplasmic fraction of the same cells expressing DMPK-CUG960 (Figure 1B) indicating that the effect was specific to nuclear fractions. To determine whether the acidic shift was due to phosphorylation, nuclear extracts were treated with alkaline phosphatase (CIAP) or treated identically but without the enzyme (mock) prior to analysis by 2D gel electrophoresis/western blotting. Nuclear CUGBP1 was acidic in untreated cells expressing DMPK-CUG960 and mock-treatment did not significantly affect the pI of CUGBP1 (Figure 1C). CIAP-treatment shifted CUGBP1 back to a more basic pI. These data indicated that expression of DMPK-CUG960 mRNA in COS M6 cells induced hyper-phosphorylation of nuclear but not cytoplasmic CUGBP1. Samples in which CUGBP1 shifted on 2D gel electrophoresis/western blotting did not exhibit a change in mobility on one dimensional gel electrophoresis (data not shown). Therefore, the hyper-phosphorylation detected here is distinct from what has been reported previously (Roberts et al., 1997).
Figure 1. DMPK-CUG960 mRNA induces hyper-phosphorylation of CUGBP1.
(A) Western blot analysis of hnRNPC (nuclear marker) or GAPDH (cytoplasmic marker) demonstrates a clean separation of nuclear and cytoplasmic extracts used for panel B. (B) Western Blot analysis of nuclear (top panel) or cytoplasmic (bottom panel) CUGBP1 in COS M6 expressing DMPK mRNA containing no repeats (DMPK-CUG0), 960 CAG repeats (DMPK-CAG960) or 960 CUG repeats (DMPK-CUG960). Proteins were separated by 2D gel electrophoresis and CUGBP1 was detected by western blot using monoclonal antibody 3B1. Results shown are representative of least three independent experiments. pH 3 to10 designates the pH range of the strips used in the experiment. (C) CIAP treatment demonstrates that the acidic shift of CUGBP1 is due to hyper-phosphorylation. Untreated, mock treated or CIAP-treated nuclear lysates were analyzed by 2D/western blotting. (D) Flag-CUGBP1 was co-expressed with DMPK-CUG960, DMPK-CUG0 mRNAs or no mRNA. Forty-eight hours post-transfection, nuclear proteins were analyzed on 2D gels and probed with HRP conjugated anti-Flag monoclonal antibodies. (E) Increased steady state levels of hyper-phosphorylated CUGBP1 in nuclear fractions of COS M6 cells expressing 960 CUG repeats. Nuclear or cytoplasmic proteins were separated by 10% SDS-PAGE and membranes were probed for CUGBP1 and GAPDH or hnRNPC as loading controls.
To determine whether exogenously expressed CUGBP1 is also modified when coexpressed with DMPK-CUG960 mRNA, an expression plasmid for Flag-tagged CUGBP1 was co-expressed with DMPK0 or DMPK-CUG960 in COS M6 cells. Nuclear fractions were analyzed by 2D gel electrophoresis and western blotting was performed using anti-Flag antibodies. As with endogenous CUGBP1, Flag-tagged CUGBP1 became hyper-phosphorylated only in cells expressing DMPK-CUG960 (Figure 1D).
CUGBP1 steady state levels are increased in DM1 cultured myoblasts as well as heart and skeletal muscle tissue cells (Dansithong et al., 2005; Ladd et al., 2005; Savkur et al., 2001; Timchenko et al., 2001). To determine whether CUGBP1 hyper-phosphorylation affects its steady state levels, we first tested whether increased phosphorylation correlated with increased steady state levels in cells transiently expressing DMPK-CUG960 mRNA. Western blot analysis revealed that CUGBP1 steady state levels were increased only in nuclear fractions but not in cytoplasmic fractions of cells expressing DMPK-CUG960 mRNA (Figure 1E). CUGBP1 levels were not increased in nuclear or cytoplasmic fractions from cells expressing DMPK0 or DMPK-CAG960, in which CUGBP1 was not hyper-phosphorylated. These results establish a strong correlation between hyper-phosphorylation and increased steady state levels.
CUGBP1 is hyper-phosphorylated in DM1 cell cultures and heart tissues
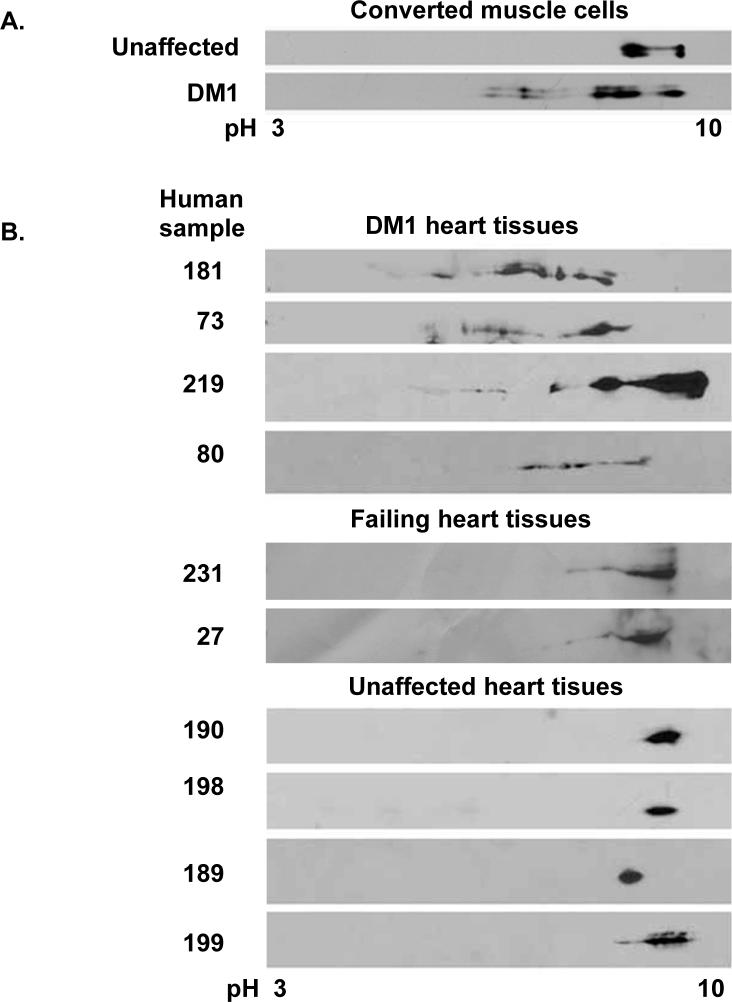
To determine whether CUGBP1 is hyper-phosphorylated in DM1 cells, we analyzed DM1 cell cultures and heart tissues. DM1 or unaffected skin fibroblasts were induced to turn on the myogenic program by retroviral-mediated expression of MyoD as described previously (Savkur et al., 2001). DM1 but not unaffected cells exhibited an embryonic pattern of cTNT splicing and elevated CUGBP1 levels as described previously (Savkur et al., 2001) (data not shown). CUGBP1 exhibited increased phosphorylation only in DM1 converted muscle cells but not in unaffected cells correlating with increased protein levels in these cells (Figure 2A).
Figure 2. CUGBP1 is hyper-phosphorylated in DM1 heart tissues and converted muscle cells.
(A) CUGBP1 hyper-phosphorylation in DM1 muscle cells. DM1 (#3989, 2000 CTG) or normal (#7492) skin fibroblasts were transduced by a retrovirus expressing MyoD and induced to undergo muscle differentiation. Whole cell extracts were analyzed by 2D/western blotting. (B) CUGBP1 hyper-phosphorylation in DM1 human heart tissues. Total cell lysates from DM1 (top panel), cardiomyopathy (middle panel) or normal (bottom panel) heart tissues were analyzed by 2D/western blotting.
We also analyzed CUGBP1 phosphorylation in human heart tissues from four individuals not affected by DM1, two individuals with non-DM1 cardiomyopathy and four individuals with DM1. All of the DM1 human heart tissues examined expressed hyper-phosphorylated forms of CUGBP1 compared to tissues from unaffected individuals and failing heart tissues (Figure 2B). We confirmed that the acidic shift of CUGBP1 was due to hyper-phosphorylation by CIAP treatment (data not shown). In addition, CUGBP1 protein levels were increased in all four DM1 heart tissues (data not shown). These results indicated that CUGBP1 is hyper-phosphorylated in DM1 cells and heart tissues and are consistent with a role for hyper-phosphorylation in increased protein levels.
CUGBP1 is hyper-phosphorylated in transgenic mice that inducibly express DMPK-CUG960 mRNA
Our lab has generated a bitransgenic mouse model for DM1 using a Cre-loxP approach to inducibly express 960 CUG repeats in the context of DMPK exon 15. Details of this model are described elsewhere (Wang et al., 2007). Bitransgenic mice contain a CTG repeat transgene (EpA960) and a heart specific, tamoxifen inducible Cre recombinase (MerCreMer or MCM). Tamoxifen administration to EpA960/MCM bitransgenic animals induces heart-specific expression of DMPK-CUG960 mRNA. These mice develop cardiac features of DM1 including arrhythmias, diastolic and systolic dysfunction, histological changes, RNA foci that colocalize with MBNL1 and characteristic alternative splicing changes. Induced mice die within two weeks after tamoxifen injection due to dilated cardiomyopathy and/or arrhythmias. Importantly, induced bitransgenic animals also exhibit elevated CUGBP1 steady state levels as is observed in DM1. RNA foci and elevated CUGBP1 was detected 6−12 hours following tamoxifen administration (Wang et al., 2007).
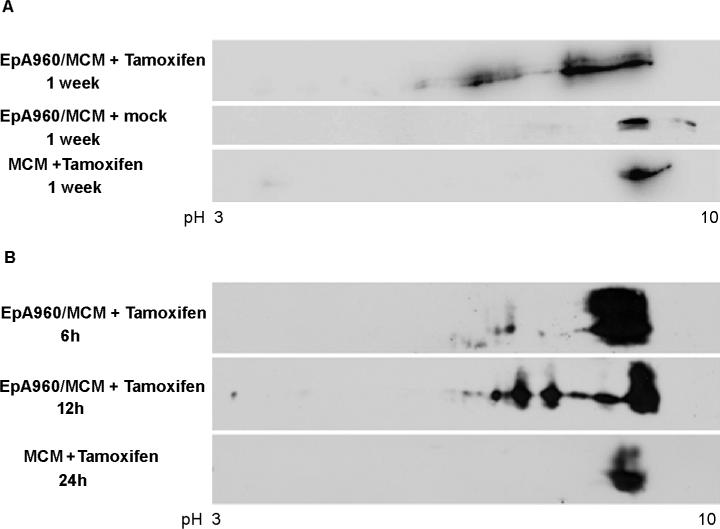
Analysis by 2D/western blotting demonstrated that CUGBP1 was consistently hyper-phosphorylated in heart tissues from tamoxifen-induced EpA960/MCM bitransgenic animals (Figure 3A). CUGBP1 was not phosphorylated in mock injected bitransgenic littermates, which did not exhibit DM1 related changes. MCM mice injected with tamoxifen also did not show a change in CUGBP1 phosphorylation, demonstrating that tamoxifen-induced Cre translocation alone did not alter CUGBP1 phosphorylation (Figure 3A). Hyper-phosphorylated CUGBP1 was clearly detectable by 12 hours by western blot suggesting that phosphorylation is an early event after repeat expression (Figure 3B). The results above indicate that CUGBP1 hyper-phosphorylation is induced by transient expression of DMPK-CUG960 mRNA in cell culture and upon induced expression of DMPK-CUG960 mRNA in a DM1 mouse model. In addition, the timing of CUGBP1 hyper-phosphorylation in the mouse model directly correlates with increased nuclear accumulation of the protein detected by immunofluorescence (Wang et al., 2007). We conclude that CUGBP1 hyper-phosphorylation is a direct effect of DMPK-CUG repeat RNA expression and induces protein stabilization.
Figure 3. CUGBP1 is hyper-phosphorylated in heart tissue from an inducible DM1 mouse model.
(A) Heart tissue from bitransgenic animals in which DMPK-CUG960 mRNA expression was induced (EpA960/MCM +Tamoxifen) or not induced (EpA960/MCM + mock) were analyzed by 2D/western blotting. MCM transgenic animals injected with tamoxifen (MCM +Tamoxifen) do not exhibit CUGBP1 hyper-phosphorylation. The results were reproduced in at least three different bitransgenic and transgenic animals. (B) Timing of CUGBP1 hyper-phosphorylation directly correlates with elevated CUGBP1 protein levels. A longer exposure was used for the 6h sample to better detect acidic shifts of CUGBP1.
PKC activation results in CUGBP1 hyper-phosphorylation and protein stabilization
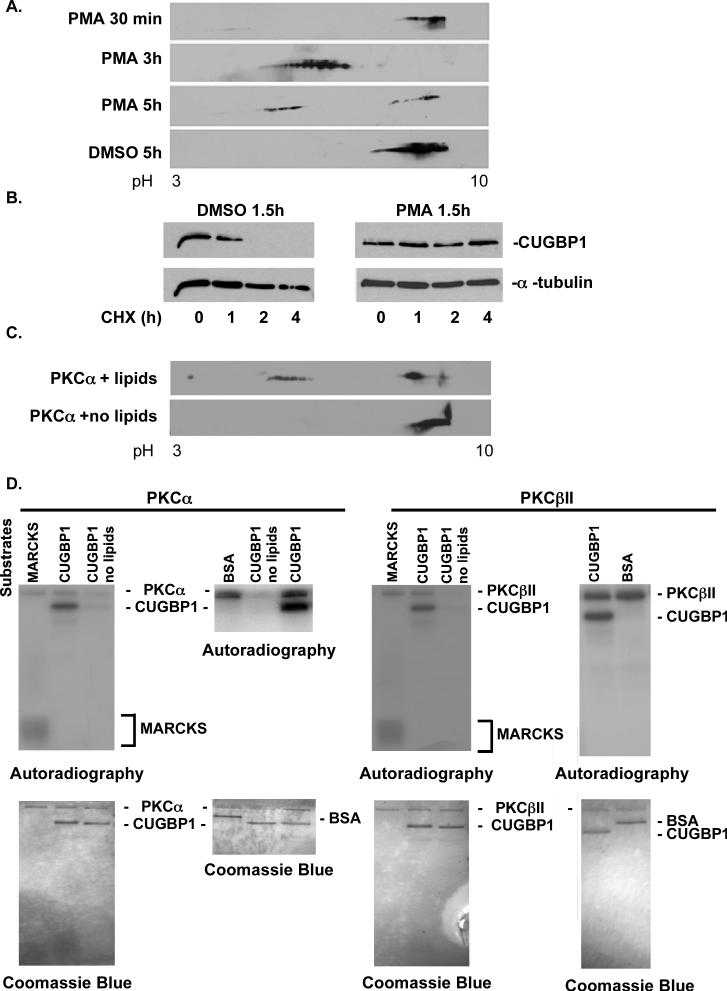
Expression of DMPK-CUG960 mRNA induces CUGBP1 hyper-phosphorylation consistent with the hypothesis that expression of DMPK-CUG960 mRNA induces a change in nuclear signaling that ultimately results in a change in CUGBP1 activity (Ranum and Cooper, 2006). We examined CUGBP1 amino acid sequence and found at least ten putative PKC, two casein kinase and two PKA phosphorylation sites. Since CUGBP1 had multiple acidic isoforms in CUG repeat expressing cells resembling multiple phosphorylation sites, we first pursued PKC with the most predicted sites. To test whether PKC is involved in CUGBP1 hyper-phosphorylation, we treated COS M6 cells with PMA to activate the PKC pathway. Following PMA treatment, total cell lysate was separated on 2D gels followed by western blot analysis. The results showed that PMA treatment resulted in CUGBP1 hyper-phosphorylation (Figure 4A). A time course following PMA addition showed increased CUGBP1 phosphorylation at 1.5 hours (data not shown), a peak at 3 hours, and a less-phosphorylated state 5 hours after treatment (Figure 4A). DMSO, the vehicle used to deliver PMA, had no effect on CUGBP1 phosphorylation (Figure 4A). These results supported a role for the PKC pathway in CUGBP1 hyper-phosphorylation.
Figure 4. PKC mediated hyper-phosphorylation of CUGBP1 increases protein stability.
(A) COS M6 cells were either treated with 30ng/ml PMA or vehicle alone (DMSO) for the indicated times. Whole cell lysates were analyzed by 2D/western blotting. (B) PMA induced hyper-phosphorylation of CUGBP1 correlates with increased half-life. COS M6 cells were treated with PMA (30ng/ml) or DMSO for 1.5 h followed by cycloheximide (10ng/ml) treatment. Cells were harvested at the indicated time points and CUGBP1 levels were determined by western blot. α-tubulin was used as a loading control. To calculate the half-life of CUGBP1, CUGBP1 and α-tubulin signal was quantified from two independent experiments using Kodak Gel Logic 2200 and Molecular Imaging Software. (C) Incubation of His-CUGBP1 with recombinant human PKCα in an in vitro kinase reaction results in an acidic shift detected by 2D/western blotting. PKCα is inactive in the absence of lipids. (D) Both human PKCα and βII directly phosphorylate His-CUGBP1 in a phospholipid dependent manner in vitro. Recombinant PKC isozymes α and βII were incubated with PKC substrate MARCKS peptide, bovine serum albumin (BSA) or His-CUGBP1 in a kinase reaction containing 32P-γ-ATP. Phosphorylated proteins were analyzed on 14% SDS-PAGE gels followed by autoradiography and Coomassie blue staining.
To test whether hyper-phosphorylation of CUGBP1 induced by PMA correlated with increased protein half-life, we measured the half-life of CUGBP1 in COS M6 cells treated with PMA or DMSO as a vehicle control. CUGBP1 hyper-phosphorylation was confirmed after PMA treatment by 2D gel/western blot analysis (data not shown). Protein synthesis was blocked by cyclohexamide and CUGBP1 half-life was determined using western blot analysis of a time course. Alpha-tubulin was used as an internal control (Carnac et al., 1998). The half-life of CUGBP1 was one hour in vehicle treated cells (Figure 4B and data not shown) and by two hours CUGBP1 was undetectable. In contrast, the half-life of hyper-phosphorylated CUGBP1 following PMA treatment was greater than 4 hours (Figure 4B). We conclude that CUGBP1 hyper-phosphorylation directly correlates with substantially longer protein half-life and increased steady state levels.
The previous results strongly suggested that CUGBP1 might be a substrate for PKC. To directly test this possibility, we performed a PKC kinase assay established previously (Ng, 2001). We focused on PKCα and βII although others are activated by PMA treatment, because it has been reported that PMA treatment causes translocation of PKCα into the nuclei of NIH 3T3 cells (Vijayan et al., 2004; Wagner et al., 2000), where phosphorylated CUGBP1 is predominant. Recombinant human PKCα was incubated with human His-CUGBP1 in a reaction mix containing ATP, calcium and lipid suspension mix, which are required for PKC activity. His-CUGBP1 was shifted to acidic forms after incubation with active PKCα as determined using 2D gels (Figure 4C). The acidic isoforms of CUGBP1 in the presence of PKCα resembled the acidic isoforms detected in PMA treated COS M6 cells (Figure 4A) providing evidence for a direct phosphorylation event. To further confirm this, we set up a kinase reaction in the presence of 32P-γ-ATP to analyze the incorporation of radiolabeled phosphate to the protein. Figure 4D shows that both recombinant human PKCα and βII specifically phosphorylates His-CUGBP1 in vitro in a Ca+2/phospholipid dependent manner, but not a BSA control. In this assay, PKCα and βII activities were confirmed by autophosphorylation of the enzymes and by phosphorylation of a known PKC target peptide MARCKS. In the absence of lipids, PKCα and βII were inactive thus did not result in autophosphorylation or phosphorylation of His-CUGBP1. Coomassie stained gels showed that similar amounts of substrates and kinases were added to each reaction. Overall, these results demonstrate that CUGBP1 is a direct target of PKCα and βII in vitro.
Expanded CUG repeat-induced hyper-phosphorylation of CUGBP1 requires PKC activity
We next sought to establish a link between the PKC pathway and CUGBP1 hyper-phosphorylation induced by DMPK-CUG960 mRNA. We first tested whether PKCα was activated in cells expressing DMPK-CUG960 mRNA. Whole cell lysates from COS M6 cells transfected with plasmids expressing DMPK-CUG960 or DMPK0 mRNA were analyzed by western blot using a polyclonal antibody that recognizes the phosphorylated (activated) forms of PKCα and PKCβII (Gysin and Imber, 1996). As a loading control, we used a monoclonal antibody that recognizes total PKCα, which also cross reacts with PKCβ. PKCα/βII was clearly activated in cells expressing DMPK-CUG960 mRNA but not in cells expressing DMPK0 mRNA (Figure 5A).
Figure 5. Induction of CUGBP1 hyper-phosphorylation by CUG repeat RNA requires PKC activity.
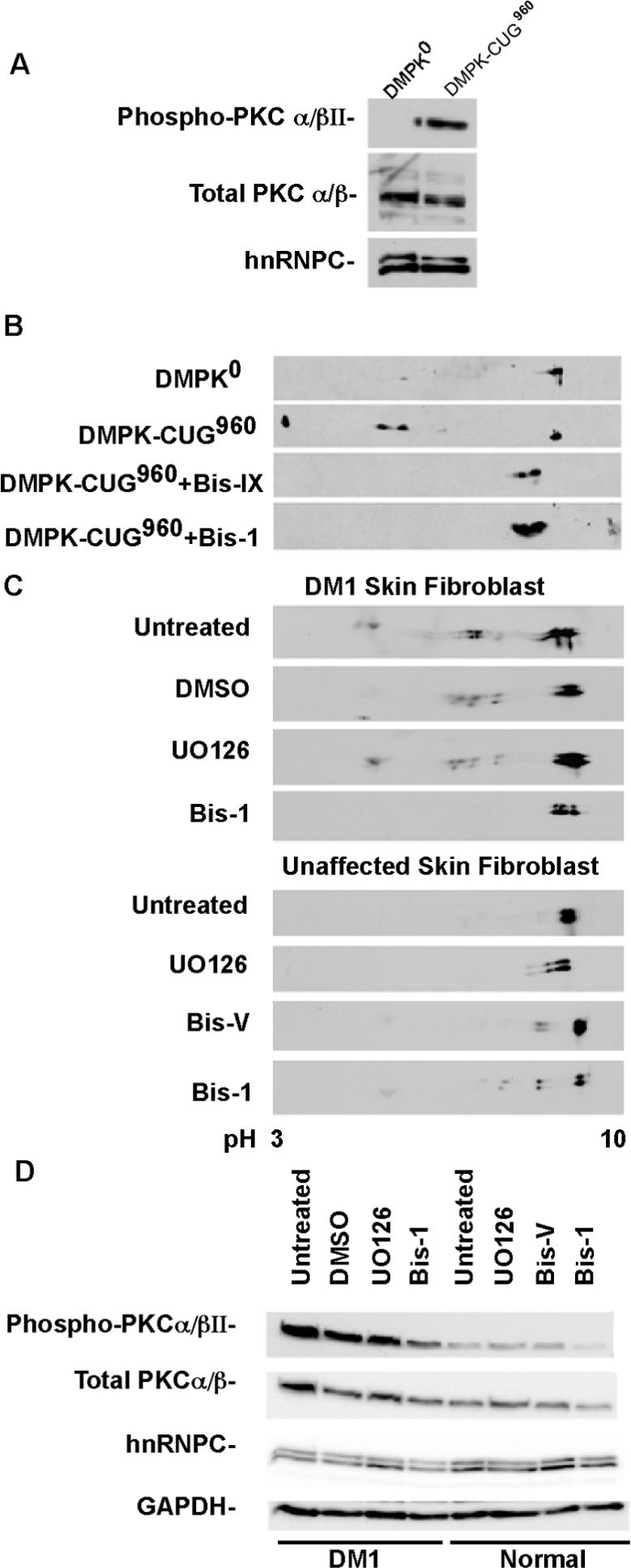
(A) Western blot analysis of total and phospho-PKCα/βII in COS M6 cells expressing DMPK-CUG0, or DMPK-CUG960. hnRNPC was used as an additional loading control. (B) PKC inhibitors blocked CUGBP1 hyper-phosphorylation in COS M6 cells expressing DMPK-CUG960 mRNA. COS M6 were transfected with DMPK-CUG0 or DMPK-CUG960. Forty-four hours post-transfection, cells were either treated with Bis-1, Bis-IX or with vehicle alone (DMSO) for 4h. CUGBP1 was analyzed by 2D/western blotting. (C) Inhibition of CUGBP1 hyper-phosphorylation in DM1 skin fibroblast (#3132, 2000 CTG) treated with Bis-1. DM1 skin fibroblast cultures were either untreated or treated with Bis-1 (PKC inhibitor), U0126 (MEK1/2 inhibitor) or DMSO for 48 h. Normal skin fibroblast (#8402) cultures were either untreated or treated with UO126, Bis-1 or Bis-V (a non-functional analogue of Bis-1) for 48 h. CUGBP1 was analyzed by 2D/western blotting. (D) Inhibition of PKC in DM1 skin fibroblast using Bis-1. The same protein samples were analyzed by 10% SDS-PAGE followed by western blot analysis using phospho-PKCα/βII, GAPDH and hnRNPC antibodies. Total PKCα/βII levels were determined after stripping the membrane probed with phospho-PKCα/βII antibody.
We next tested whether PKC activity was required for CUGBP1 hyper-phosphorylation in response to expression of DMPK-CUG960 mRNA. Cells transfected with DMPK-CUG960 were treated with two specific inhibitors of PKC, Bis-IX or Bis-1. Both inhibitors completely blocked hyper-phosphorylation of CUGBP1 induced by DMPK-CUG960 mRNA (Figure 5B) indicating that PKC activity is required for the trans effect of repeat-containing RNA on CUGBP1 hyper-phosphorylation.
We wanted to confirm whether PKC activity was required for CUGBP1 hyper-phosphorylation in DM1 cultured cells. CUGBP1 is hyper-phosphorylated in DM1 skin fibroblasts (Figure 5C) as observed in converted myoblasts above (Figure 2A). The hyper-phosphorylated state of CUGBP1 was lost in DM1 fibroblasts following treatment for 48 hours with Bis-1. This effect was specific because neither vehicle alone (DMSO) nor the unrelated MEK1/2 kinase inhibitor (U0126) inhibited CUGBP1 hyper-phosphorylation. Prolonged treatment with Bis-1 caused a slightly acidic shift of CUGBP1 in normal fibroblasts (Figure 5C) suggesting that prolonged inactivation of PKC triggers phosphorylation of CUGBP1 by additional pathways.
Western blot analysis for phosphorylated PKCα/βII clearly demonstrated that PKCα/βII was activated in DM1 skin fibroblasts and that activation was reduced by Bis-1 treatment (Figure 5D). These results demonstrate that PKCα/βII is activated in DM1 fibroblasts compared to fibroblasts from unaffected individuals and that PKCα/βII activation directly correlates with CUGBP1 hyper-phosphorylation in these cells (Figure 5D). Taken together the results above indicate that activation of PKC isozymes is required for CUGBP1 phosphorylation in DM1 skin fibroblasts.
PKCα/βII is activated in heart tissues from the DM1 mouse model and individuals with DM1
The cell culture model indicated a role for the PKC pathway in hyper-phosphorylation of CUGBP1 in response to DMPK-CUG960 mRNA expression. To determine whether activation of PKC pathway was a feature of DM1, we assayed for PKCα/βII activation in heart tissues from the DM1 mouse model and individuals with DM1. As shown in Figure 6A, PKCα/βII phosphorylation was induced in all of the EpA960/MCM mice in which DMPK-CUG960 mRNA expression was induced with tamoxifen but not in the mock injected EpA960/MCM (data not shown) or tamoxifen injected MCM littermates. PKCα/βII was activated by 6 hours after tamoxifen injection, the earliest time examined (Figure 6B). Analysis of CUGBP1 steady state levels demonstrated an increase in CUGBP1 in all mice that expressed DMPK-CUG960 mRNA and activated PKCα/βII consistent with the finding that CUGBP1 levels are elevated in DM1 cells and tissues (Dansithong et al., 2005; Savkur et al., 2001; Timchenko et al., 2001). PKCα/βII was also activated in embryonic day 16 heart tissues correlating with CUGBP1 hyper-phosphorylation (see below) and increased steady state levels in embryonic tissues (Figure 6A). These results indicated that PKCα/βII is activated soon after induction of DMPK-CUG960 mRNA in mouse heart.
Figure 6. PKCα is activated in DM1.
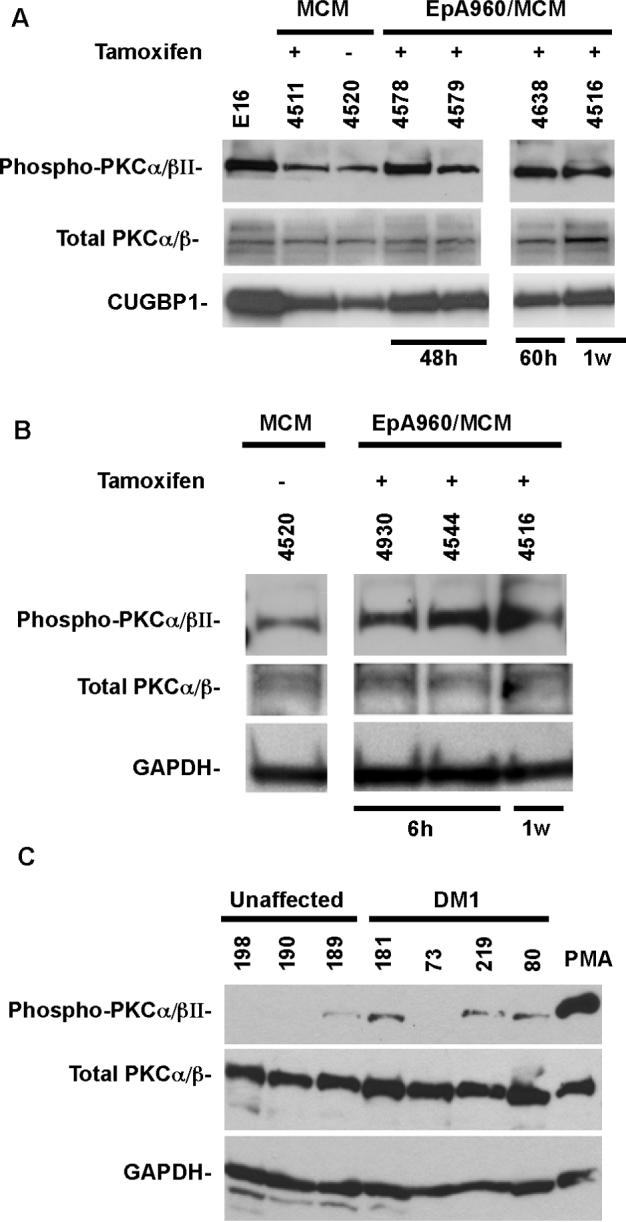
(A-B) PKCα/βII activation in a DM1 mouse model inducibly expressing 960 CUG repeats in heart. Western blot analysis of total and phospho-PKCα/βII and CUGBP1 in normal embryonic day 16 (e16) heart, MCM and EpA960/MCM mice heart tissues. Tamoxifen administration is indicated as (+) or (–). Heart tissues were analyzed at indicated times after tamoxifen injection. GAPDH was used as an additional loading control. (C) PKCα/βII activation in DM1 heart tissues that were analyzed for CUGBP1 phosphorylation in Figure 2. Sample 199 (not shown) failed to show a signal for total PKCα/βII and showed a weak signal for GAPDH. Total and phospho-PKCα/βII levels were determined as described above.
To determine whether PKCα/βII was activated in DM1 patient samples, we tested the heart tissues analyzed for CUGBP1 phosphorylation in Figure 2B. Lysate from PMA treated COS M6 cells was used as a positive control for PKC activation. Three out of four DM1 patient heart tissues displayed PKCα/βII activation (Figure 6C). The absence of signal in one of the DM1 samples might be due to differences in handling and processing of tissue after biopsy as well as individual to individual variation. Only one of the three normal heart tissues exhibited slight PKCα/βII activation. However, the activation observed in this unaffected individual was not as robust as the activation in DM1 subjects.
CUGBP1 hyper-phosphorylation correlates with abundance during striated muscle development
To examine a potential relationship between CUGBP1 phosphorylation and regulation of protein steady state levels during development, we examined endogenous CUGBP1 in hearts from embryonic day 17 (e17), newborn and adult mice and skeletal muscle from newborn and adult mice. We found that CUGBP1 was more acidic in both newborn and e17 heart tissues as well as in newborn skeletal muscle tissues compared to heart or skeletal muscle from adults (Figure 7A and 7B). The acidic shift of CUGBP1 in newborn heart was due to hyper-phosphorylation determined by CIAP treatment (Figure 7C). We conclude that a developmental change in the phosphorylation state of CUGBP1 in heart and skeletal muscle directly correlates with its steady state protein levels.
Figure 7. CUGBP1 is hyper-phosphorylated in newborn and embryonic heart and skeletal muscle tissues.
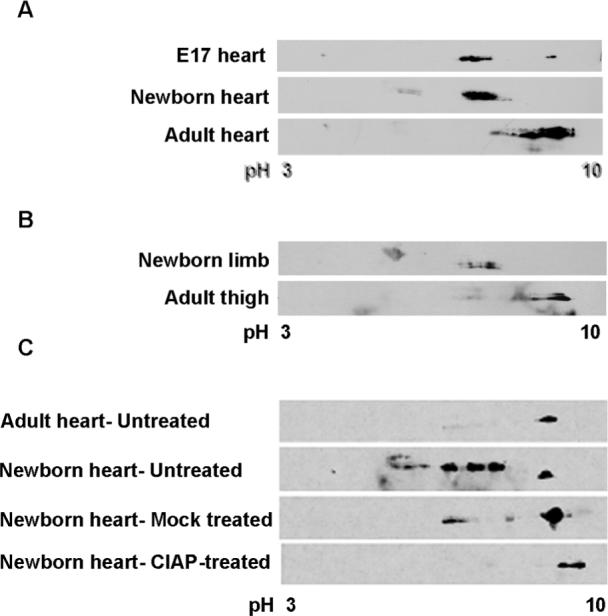
CUGBP1 phosphorylation was analyzed on 2D gels (A) during heart development (e17, newborn and 6 months old adult mice), and (B) during skeletal muscle development (newborn and 6 months old adult mice) by western blot using HRP conjugated CUGBP1 monoclonal antibody. Seventy-five micrograms of total protein from e17 and newborn tissues and one hundred fifty micrograms of adult heart tissues were analyzed. Representative results from at least three independent experiments are shown. Note that e17 heart samples were a pool of eight mice hearts. (C) CUGBP1 is hyper-phosphorylated in newborn heart. Untreated, mock treated or CIAP-treated newborn heart tissues were analyzed by 2D/western blotting.
Discussion
Here we demonstrate that expression of DMPK-CUG repeat RNA induces hyper-phosphorylation of CUGBP1, which results in increased protein half-life and steady state levels. CUGBP1 was demonstrated to be hyper-phosphorylated in four different experimental systems including COS M6 cells expressing DMPK-CUG960 RNA, DM1 cell cultures, DM1 tissues, and an inducible DM1 mouse model. We also demonstrated that CUGBP1 hyper-phosphorylation in response to transient expression of DMPK-CUG960 RNA and in DM1 cultures requires PKC activity and that activation of PKC using a phorbol ester causes CUGBP1 hyper-phosphorylation and significantly increases protein half-life. CUGBP1 was directly phosphorylated by both PKC α and βII isozymes in an in vitro kinase assay. PKCα/βII was found to be activated in cells expressing DMPK-CUG960 RNA, DM1 cell cultures, DM1 tissues, and the inducible DM1 mouse model. Furthermore, a time course of molecular events following induction of DMPK-CUG960 RNA in mouse heart demonstrated that PKCα/βII activation, CUGBP1 hyper-phosphorylation, and elevation of CUGBP1 steady state levels occurred within six hours following induction of RNA expression. These results are consistent with a cause-effect relationship between expression of DMPK-CUG960 RNA, CUGBP1 hyper-phosphorylation, and increased CUGBP1 steady state levels. The findings from this paper provides a model for DM1 in which DMPK-CUG960 RNA triggers a signaling event that leads to PKCα/βII activation. Increased PKCα/βII activity causes hyper-phosphorylation of CUGBP1, most likely by a direct phosphorylation event. This phosphorylation event prolongs the half-life and increases the steady state levels of CUGBP1. Elevated CUGBP1 activity potentially leads to abnormalities in adult heart tissue by altering splicing of pre-mRNA targets and translation of target mRNAs.
CUGBP1 was previously shown to be a phosphoprotein and the phosphorylation state was found to be altered in DM1 heart tissue (Roberts et al., 1997). In this previous study, analysis on one dimensional gels indicated nuclear accumulation of a hypo-phosphorylated isoform in DM1 tissues (Roberts et al., 1997). This differs from our results in which expression of DMPK-CUG960 mRNA induced hyper-phosphorylation exclusively of nuclear CUGBP1 in cultured cells. In addition, we observed hyper-phosphorylation of CUGBP1 in DM1 tissue samples, cultured DM1 cells as well as within 6 hours following induction of DMPK-CUG960 mRNA in heart tissues from an DM1 inducible mouse model (Wang et al., 2007). The differences in CUGBP1 mobility assayed on a 1D gel in the previous study do not represent the phosphorylation events identified in this report by the 2D analysis because the phosphorylation changes induced by transient expression of DMPK-CUG960 mRNA that were observed by 2D gel analysis were not apparent on 1D gels (data not shown).
Identification of multiple putative PKC phosphorylation sites on CUGBP1 led us to investigate the role of PKC in CUGBP1 phosphorylation. The PKC pathway is involved in many cellular processes such as growth, apoptosis, and differentiation (Musashi et al., 2000). There are twelve PKC isozymes, all of which are serine/threonine kinases. Activation of these isozymes requires subsequent phosphorylation, lipid binding and translocation to membranes (Musashi et al., 2000). Using activators and inhibitors of the PKC pathway, we identified PKC as required for CUGBP1 hyper-phosphorylation. We also demonstrated that recombinant PKCα and βII directly phosphorylate His-CUGBP1 in an in vitro kinase reaction. Several PKC isozymes translocate to the nucleus upon activation and regulate nuclear events such as transcription, splicing and mRNA stability via phosphorylation of nuclear targets such as lamin B and histone H1 (Fields et al., 1988; Martelli et al., 2006; Martelli et al., 2003; Omri et al., 1987). It is also possible that CUGBP1 is phosphorylated in the cytoplasm and is efficiently translocated to the nucleus.
We found that that CUGBP1 hyper-phosphorylation mediated both by expression of expanded CUG repeats and PKC activation by phorbol esters leads to increased protein half-life. This is consistent with a previous report showed that CUGBP1 half-life was increased in COS7 cells expressing 170 CUG repeats (Timchenko et al., 2001). Interestingly, the neuron specific embryonic lethal abnormal vision (nELAV) proteins, RNA binding proteins that are structurally similar to CUGBP1, are phosphorylated by PKCα after PMA treatment. PKC mediated phosphorylation increased the steady state levels of these proteins in rat hippocampus (Pascale et al., 2005). It is possible that PKCα-mediated phosphorylation of CUGBP1 and related proteins could be a common mechanism for regulation of protein levels.
A key molecular feature of DM is the expression of embryonic alternative splicing patterns in adult tissues. For the ClC1 and IR genes, the failure of the embryonic isoforms to fulfill the functional requirements of adult tissues results in myotonia and insulin resistance, respectively (Charlet-B. et al., 2002; Mankodi et al., 2002; Savkur et al., 2001). It is of interest that the first two proteins identified as CUG repeat binding proteins, CUGBP1 and MBNL1, are antagonistic regulators of alternative splicing and that both proteins normally regulate alternative splicing transitions during development (Ho et al., 2004; Ladd et al., 2005; Lin et al., 2006). Normal modulation of MBNL1 activity during development is due to a postnatal transition from predominantly cytoplasmic to predominantly nuclear localization in skeletal muscle (Lin et al., 2006). CUGBP1 protein expression is tightly down-regulated during normal post-natal development of heart and skeletal muscle (Ladd et al., 2005; Lin et al., 2006). However, CUGBP1 mRNA levels do not exhibit a corresponding decrease suggesting that protein steady state levels are regulated at the level of translation or protein stability (Ladd et al., 2005; Lin et al., 2006). We show a strong correlation between CUGBP1 hyper-phosphorylation, elevated steady state levels of CUGBP1 protein, and PKCα/βII activation in early developmental stages as well as DM1 tissues. In support of a role for PKC in modulating CUGBP1 levels, PKCα and βII levels are down regulated during heart development (Hamplova et al., 2005; Schreiber et al., 2001). These results strongly support a model in which DMPK-CUG960 mRNA expression disrupts the normal developmental regulation of CUGBP1 steady state levels mediated via a PKC pathway. These findings provide potential therapy options to reduce abnormal CUGBP1 levels to ameliorate DM1 phenotype.
Experimental Procedure
Plasmids
The human cTNT minigene was described previously (Philips et al., 1998). DMPK-CUG0, plasmids expressing DMPK exons 11 to 15 with no repeats (DMPK-CUG0), with 960 interrupted CAG repeats (DMPK-CAG960) and with 960 interrupted CTG repeats (DMPK-CTG960) were described previously (Ho et al., 2005b; Philips et al., 1998).
Tissue Culture Cells and conversion of skin fibroblasts to muscle cells and human tissues
COS M6 cells were maintained in Dulbecco's modified Eagle's medium (DMEM, Gibco) containing 10% fetal bovine serum (FBS), 1% penicillin-streptomycin (Gibco), 1% L-Glutamine (Gibco) and 1% fungizone (Gibco) at 370 C in 5% CO2.
DM1 (#3989) with 2000 CTG repeats and unaffected (#7492) skin fibroblasts were purchased from Coriell Cell Repositories (Camden, NJ). Fibroblast cells were maintained in MEM supplemented with 15% FBS, 2% essential amino acids (Gibco), 1% non-essential amino acids, 1% L-glutamine, 1% penicillin-streptomycin (Gibco) and 0.92% of 1M NaOH at 370 C in 5% CO2. Cells were split according to the protocol by Coriell Cell Repositories. To convert these skin fibroblasts to muscle cells, 20% confluent skin fibroblasts were infected with an amphotrophic retrovirus expressing MyoD for 3.5 hours (Weintraub et al., 1989). After three days, cells were selected in complete media containing 0.8mg/ml G418 (Gibco) and differentiated for five days as described previously (Savkur et al., 2001). The differentiation status of both normal and DM1 muscle cells were similar assessed by immunofluorescence using myosin heavy chain specific monoclonal antibody (MF20 from Developmental Studies Hybridoma Bank, Iowa city, Iowa) (data not shown). RT-PCR was performed to determine the splicing changes of endogenous cTNT and IR in these cells as previously described (data not shown) (Philips et al., 1998; Savkur et al., 2001).
COS M6 cells (1.5 × 105) were plated into a 60mm dish. Twenty-four hours after plating, cells were transfected with 1μg of repeat containing plasmids and 1μg of carrier DNA (sp72) to make total DNA concentration to 2μg per dish using Fugene 6 (Roche) according to the manufacturer's protocol. Cells were harvested 48h post-transfection unless otherwise stated in figure legends.
COS M6 cells were either treated with 30ng/ml PMA or DMSO for indicated time intervals. Cells were scraped in 2D urea buffer followed by sonication and total cell lysates were analyzed on 2D gels followed by western blot using CUGBP1 monoclonal antibody.
COS M6 cells were treated with cycloheximide (10μg/ml), then harvested at different time points after cycloheximide treatment. Protein lysates were analyzed on SDS-PAGE followed by CUGBP1 western blotting.
Nuclear-cytoplasmic fractions were isolated as described previously (Ladd et al., 2005). Nuclei pellets were resuspended in 2D urea buffer (7M urea, 2M thiourea, 20mM Tris pH 7.5, 4% CHAPS) and cytoplasmic fractions were acetone precipitated at –800C for 2hr and pelleted down for 5min at 13,000 rpm on benchtop centrifuge and resuspended either in 2D urea buffer before loading on 2D gels or in 4x Laemnli buffer to analyze on 1D gels. Human heart tissue samples were obtained from NDRI, Dr. Charles Thornton, Dr. Tetsuo Ashizawa, and CHTN University of Pennsylvania.
Sample preparation
Human and mouse heart tissues were immediately frozen in liquid nitrogen and grinded using a mortal and a pestle. The powderized tissue was solubilized in 2D urea buffer and was sonicated gently three times. The lysate was cleared after centrifugation for 5min at 13,000 rpm on a bench top centrifuge. Protein concentrations of the cleared lysates were determined by Bradford assay (BioRad).
For cultured cells, cells were washed with 1xPBS. After removal of 1xPBS, cells were scraped in 2D urea buffer or in 4x Laemnli buffer. Lysates were sonicated and cleared by centrifugation at 13,000 rpm for 5min followed by protein assay.
CIAP treatment
Powderized tissues were resuspended in lysis buffer containing phosphatase inhibitors (5mM NaCl, 20mM HEPES, pH 7.5, 6mM pNPP, 1mM sodium pervanadate and complete protease inhibitor cocktail and 0.4% NP-40). CIAP-treated lysates were incubated at RT for 15min with 1 unit of CIAP/25μg of nuclear protein. Mock treated lysates were incubated at room temperature for 15 minutes with CIAP buffer. Untreated controls were kept on ice for 15min. Each lysate (40μg) were separated on 2D gel electrophoresis and the pI of CUGBP1 was analyzed by western blot using CUGBP1 specific monoclonal antibody. For cultured cells, cells were scraped in the same lysis buffer described above and CIAP treatments were performed as described above.
2D gel analysis
First dimension of 2D gel analysis was performed on Ettan IPGphor Isoelectric Focusing System (GE Healthcare) according to the manufacturer's protocol. Briefly, protein lysates were loaded onto the ceramic strip holders (GE Healthcare) and the frozen immobilized IPG strips (BioRad) were layered on the samples for rehydration followed by isoelectric focusing (IEF) overnight at 30°C. Then, IPG strips were loaded on precast 10% IPG- 1 well resolving gels (BioRad) with a prestained marker (BioRad) and gels were run for 1.5h at 185V. The proteins were transferred to PVDF membrane followed by western blotting.
Western Blot and antibodies
CUGBP1 was detected using CUGBP1 monoclonal antibody (3B1) (Upstate, 1 to 2500). Incubations were held overnight at 40C in 4% blotto with 0.1% Tween 20. Anti-mouse IgG-HRP (Jackson, 1 to 2500) was used as a secondary antibody in 4% blotto. Membranes were washed in PBS + 0.5% Tween 20 for 15min three times between incubations at room temperature. HnRNPC (4F4) monoclonal antibody (Abcam, 1 to 10,000), and GAPDH monoclonal antibody (Biogenesis, 1 to 10,000) were used as loading controls.
For detection of CUGBP1 in tissues, CUGBP1 monoclonal antibody was conjugated to horseradish peroxidase (HRP) to avoid detection of mouse IgG. The conjugation of to CUGBP1 monoclonal antibody was performed using EZ-Link Plus Activated Peroxidase kit (Pierce, Rockford, IL) and purified using the FreeZyme Conjugate Purification Kit (Pierce, Rockford, IL) (Pierce, Rockford, IL) as described previously (Ladd et al., 2005). CUGBP1-HRP antibody was incubated overnight at 40C at a dilution of 1 to 100.
In vitro kinase assay with human recombinant PKC kinase and His-CUGBP1
The PKC kinase assay was assembled according to the protocol described previously (Ng, 2001). Briefly, the assay was set up using 400ng of GST-tagged PKCα or βII (Cell Signaling) in a 35μl reaction containing a lipid suspension mix (50μg phosphatidylserine, 50μg phosphatidylinositol 4,5 bi-phosphate and 0.5μg PMA), 0.8mM ATP mixed with 1μCi 32P-γ-ATP, 6mM CaCl2, 100mM MgCl2 and the following substrates: 80ng of MARCKS peptide (Cell Signaling) a positive control as a PKC substrate, 800ng BSA or 400ng of recombinant human His-CUGBP1. The enzyme to substrate molar ratio was 1 to 2. The reaction was incubated for 40min at 300 C and stopped by addition of 4x Laemmli buffer. Half of the reaction volume was loaded on 14% SDS-PAGE. Phosphorylated proteins were analyzed by autoradiography. The same gel was stained by Coomassie blue and an image was captured using the Kodak Gel Logic 2200.
Acknowledgements
We thank D. Engler and R. Matsunami for help establishing 2D gel electrophoresis and analysis of the data and The Comparative Human Tissue Network (CHTN), The National Development and Resource Institute (NDRI) and Drs. C. Thornton and T. Ashizawa for tissue samples. We also thank R.T. Javier and J.M. Rosen for critically reading the manuscript. This work was supported by NIH NRSA (1F32AR052630-01) to N.M.K and by NIH (R01AR45653 and R01HL45565) and the Muscular Dystrophy Association to T.A.C.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Cardani R, Mancinelli E, Rotondo G, Sansone V, Meola G. Muscleblind-like protein 1 nuclear sequestration is a molecular pathology marker of DM1 and DM2. Eur J Histochem. 2006;50:177–182. [PubMed] [Google Scholar]
- Carnac G, Primig M, Kitzmann M, Chafey P, Tuil D, Lamb N, Fernandez A. RhoA GTPase and serum response factor control selectively the expression of MyoD without affecting Myf5 in mouse myoblasts. Mol Biol Cell. 1998;9:1891–1902. doi: 10.1091/mbc.9.7.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlet-B. N, Savkur RS, Singh G, Philips AV, Grice EA, Cooper TA. Loss of the muscle-specific chloride channel in type 1 myotonic dystrophy due to misregulated alternative splicing. Mol Cell. 2002;10:45–53. doi: 10.1016/s1097-2765(02)00572-5. [DOI] [PubMed] [Google Scholar]
- Dansithong W, Paul S, Comai L, Reddy S. MBNL1 is the primary determinant of focus formation and aberrant insulin receptor splicing in DM1. J Biol Chem. 2005;280:5773–5780. doi: 10.1074/jbc.M410781200. [DOI] [PubMed] [Google Scholar]
- Fardaei M, Larkin K, Brook JD, Hamshere MG. In vivo co-localisation of MBNL protein with DMPK expanded-repeat transcripts. Nucleic Acids Res. 2001;29:2766–2771. doi: 10.1093/nar/29.13.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fardaei M, Rogers MT, Thorpe HM, Larkin K, Hamshere MG, Harper PS, Brook JD. Three proteins, MBNL, MBLL and MBXL, co-localize in vivo with nuclear foci of expanded-repeat transcripts in DM1 and DM2 cells. Hum Mol Genet. 2002;11:805–814. doi: 10.1093/hmg/11.7.805. [DOI] [PubMed] [Google Scholar]
- Fields AP, Pettit GR, May WS. Phosphorylation of lamin B at the nuclear membrane by activated protein kinase C. J Biol Chem. 1988;263:8253–8260. [PubMed] [Google Scholar]
- Gysin S, Imber R. Replacement of Ser657 of protein kinase C-alpha by alanine leads to premature down regulation after phorbol-ester-induced translocation to the membrane. Eur J Biochem. 1996;240:747–750. doi: 10.1111/j.1432-1033.1996.0747h.x. [DOI] [PubMed] [Google Scholar]
- Hamplova B, Novakova O, Tvrzicka E, Kolar F, Novak F. Protein kinase C activity and isoform expression during early postnatal development of rat myocardium. Cell Biochem Biophys. 2005;43:105–117. doi: 10.1385/CBB:43:1:105. [DOI] [PubMed] [Google Scholar]
- Harper PS. Myotonic Dystrophy. Third edn Vol. 37. W.B. Saunders; London: 2001. [Google Scholar]
- Ho TH, Bundman D, Armstrong DL, Cooper TA. Transgenic mice expressing CUG-BP1 reproduce splicing mis-regulation observed in myotonic dystrophy. Hum Mol Genet. 2005a;14:1539–1547. doi: 10.1093/hmg/ddi162. [DOI] [PubMed] [Google Scholar]
- Ho TH, Charlet-B. N, Poulos MG, Singh G, Swanson MS, Cooper TA. Muscleblind proteins regulate alternative splicing. EMBO J. 2004;23:3103–3112. doi: 10.1038/sj.emboj.7600300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho TH, Savkur R, Poulos MG, Mancini MA, Swanson MS, Cooper TA. Co-localization of muscleblind with RNA foci is separable from mis-regulation of alternative splicing in myotonic dystrophy. J Cell Sci. 2005b;118:2923–2933. doi: 10.1242/jcs.02404. [DOI] [PubMed] [Google Scholar]
- Jiang H, Mankodi A, Swanson MS, Moxley RT, Thornton CA. Myotonic dystrophy type 1 is associated with nuclear foci of mutant RNA, sequestration of muscleblind proteins and deregulated alternative splicing in neurons. Hum Mol Genet. 2004;13:3079–3088. doi: 10.1093/hmg/ddh327. [DOI] [PubMed] [Google Scholar]
- Kanadia RN, Johnstone KA, Mankodi A, Lungu C, Thornton CA, Esson D, Timmers AM, Hauswirth WW, Swanson MS. A muscleblind knockout model for myotonic dystrophy. Science. 2003;302:1978–1980. doi: 10.1126/science.1088583. [DOI] [PubMed] [Google Scholar]
- Kuyumcu-Martinez NM, Cooper TA. Misregulation of alternative splicing causes pathogenesis in myotonic dystrophy. Prog Mol Subcell Biol. 2006;44:133–159. doi: 10.1007/978-3-540-34449-0_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladd AN, Stenberg MG, Swanson MS, Cooper TA. Dynamic balance between activation and repression regulates pre-mRNA alternative splicing during heart development. Dev Dyn. 2005;233:783–793. doi: 10.1002/dvdy.20382. [DOI] [PubMed] [Google Scholar]
- Lin X, Miller JW, Mankodi A, Kanadia RN, Yuan Y, Moxley RT, Swanson MS, Thornton CA. Failure of MBNL1-dependent post-natal splicing transitions in myotonic dystrophy. Hum Mol Genet. 2006;15:2087–2097. doi: 10.1093/hmg/ddl132. [DOI] [PubMed] [Google Scholar]
- Lueck JD, Lungu C, Mankodi A, Osborne R, Welle S, Dirksen RT, Thornton CA. Chloride channelopathy in myotonic dystrophy resulting from loss of post-transcriptional regulation for CLCN1. Am J Physiol Cell Physiol. 2007;292:C1245–1247. doi: 10.1152/ajpcell.00336.2006. [DOI] [PubMed] [Google Scholar]
- Mahadevan MS, Yadava RS, Yu Q, Balijepalli S, Frenzel-McCardell CD, Bourne TD, Phillips LH. Reversible model of RNA toxicity and cardiac conduction defects in myotonic dystrophy. Nat Genet. 2006;38:1066–1070. doi: 10.1038/ng1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankodi A, Lin X, Blaxall BC, Swanson MS, Thornton CA. Nuclear RNA foci in the heart in myotonic dystrophy. Circ Res. 2005;97:1152–1155. doi: 10.1161/01.RES.0000193598.89753.e3. [DOI] [PubMed] [Google Scholar]
- Mankodi A, Takahashi MP, Jiang H, Beck CL, Bowers WJ, Moxley RT, Cannon SC, Thornton CA. Expanded CUG repeats trigger aberrant splicing of ClC-1 chloride channel pre-mRNA and hyperexcitability of skeletal muscle in myotonic dystrophy. Mol Cell. 2002;10:35–44. doi: 10.1016/s1097-2765(02)00563-4. [DOI] [PubMed] [Google Scholar]
- Martelli AM, Evangelisti C, Nyakern M, Manzoli FA. Nuclear protein kinase C. Biochim Biophys Acta. 2006;1761:542–551. doi: 10.1016/j.bbalip.2006.02.009. [DOI] [PubMed] [Google Scholar]
- Martelli AM, Faenza I, Billi AM, Fala F, Cocco L, Manzoli L. Nuclear protein kinase C isoforms: key players in multiple cell functions? Histol Histopathol. 2003;18:1301–1312. doi: 10.14670/HH-18.1301. [DOI] [PubMed] [Google Scholar]
- Musashi M, Ota S, Shiroshita N. The role of protein kinase C isoforms in cell proliferation and apoptosis. Int J Hematol. 2000;72:12–19. [PubMed] [Google Scholar]
- Ng T, Parsons M, Hughes WE, Monypenny J, Zicha D, Gautreau A, Arpin M, Gschmeissner S, Verveer PJ, Bastiaens PI, Parker PJ. Ezrin is a donwstream effector of trafficking PKC-integrin complexes involved in the control of cell motility. EMBO J. 2001;20:2723–2741. doi: 10.1093/emboj/20.11.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omri B, Breton MF, Pavlovic-Hournac M. Characteristics of thyroid protein kinase C. Different Ca2 requirement for the phosphorylation of endogenous proteins and of H1 histone. Eur J Biochem. 1987;165:83–90. doi: 10.1111/j.1432-1033.1987.tb11197.x. [DOI] [PubMed] [Google Scholar]
- Pascale A, Amadio M, Scapagnini G, Lanni C, Racchi M, Provenzani A, Govoni S, Alkon DL, Quattrone A. Neuronal ELAV proteins enhance mRNA stability by a PKCalpha-dependent pathway. Proc Natl Acad Sci U S A. 2005;102:12065–12070. doi: 10.1073/pnas.0504702102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philips AV, Timchenko LT, Cooper TA. Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science. 1998;280:737–741. doi: 10.1126/science.280.5364.737. [DOI] [PubMed] [Google Scholar]
- Ranum LP, Cooper TA. RNA-Mediated Neuromuscular Disorders. Annu Rev Neurosci. 2006;38:758–769. doi: 10.1146/annurev.neuro.29.051605.113014. [DOI] [PubMed] [Google Scholar]
- Roberts R, Timchenko NA, Miller JW, Reddy S, Caskey CT, Swanson MS, Timchenko LT. Altered phosphorylation and intracellular distribution of a (CUG)n triplet repeat RNA-binding protein in patients with myotonic dystrophy and in myotonin protein kinase knockout mice. Proc Natl Acad Sci. 1997;94:13221–13226. doi: 10.1073/pnas.94.24.13221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savkur RS, Philips AV, Cooper TA. Aberrant regulation of insulin receptor alternative splicing is associated with insulin resistance in myotonic dystrophy. Nat. Gen. 2001;29:40–47. doi: 10.1038/ng704. [DOI] [PubMed] [Google Scholar]
- Schreiber KL, Paquet L, Allen BG, Rindt H. Protein kinase C isoform expression and activity in the mouse heart. Am J Physiol Heart Circ Physiol. 2001;281:H2062–2071. doi: 10.1152/ajpheart.2001.281.5.H2062. [DOI] [PubMed] [Google Scholar]
- Timchenko LT, Timchenko NA, Caskey CT, Roberts R. Novel proteins with binding specificity for DNA CTG repeats and RNA CUG repeats -implications for myotonic dystrophy. Hum Mol Genet. 1996;5:115–121. doi: 10.1093/hmg/5.1.115. [DOI] [PubMed] [Google Scholar]
- Timchenko NA, Cai ZJ, Welm AL, Reddy S, Ashizawa T, Timchenko LT. RNA CUG repeats sequester CUGBP1 and alter protein levels and activity of CUGBP1. J Biol Chem. 2001;276:7820–7826. doi: 10.1074/jbc.M005960200. [DOI] [PubMed] [Google Scholar]
- Timchenko NA, Patel R, Iakova P, Cai ZJ, Quan L, Timchenko LT. Overexpression of CUG triplet repeat-binding protein, CUGBP1, in mice inhibits myogenesis. J Biol Chem. 2004;279:13129–13139. doi: 10.1074/jbc.M312923200. [DOI] [PubMed] [Google Scholar]
- Vijayan K, Szotek EL, Martin JL, Samarel AM. Protein kinase C-alpha-induced hypertrophy of neonatal rat ventricular myocytes. Am J Physiol Heart Circ Physiol. 2004;287:H2777–2789. doi: 10.1152/ajpheart.00171.2004. [DOI] [PubMed] [Google Scholar]
- Wagner S, Harteneck C, Hucho F, Buchner K. Analysis of the subcellular distribution of protein kinase Calpha using PKC-GFP fusion proteins. Exp Cell Res. 2000;258:204–214. doi: 10.1006/excr.2000.4925. [DOI] [PubMed] [Google Scholar]
- Wang GS, Kearney DL, De Biasi M, Taffet G, Cooper T. Elevated CUGBP1 is an early event in an inducible heart-specific mouse model of myotonic dystrophy. J Clin Invest. 2007 doi: 10.1172/JCI32308. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H, Tapscott SJ, Davis RL, Thayer MJ, Adam MA, Lassar AB, Miller AD. Activation of muscle-specific genes in pigment, nerve, fat, liver, and fibroblast cell lines by forced expression of MyoD. Proc. Natl. Acad. Sci. USA. 1989;86:5434–5438. doi: 10.1073/pnas.86.14.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]