SUMMARY
IF3 has a fidelity function in the initiation of translation, inducing the dissociation of fMet-tRNAfMet from 30S initiation complexes (30SIC) containing a non-canonical initiation triplet (e.g., AUU) in place of a canonical initiation triplet (e.g., AUG). IF2 plays a complementary role, selectively promoting initiator tRNA binding to the ribosome. Here we use parallel rapid kinetics measurements of GTP hydrolysis, Pi release, light scattering, and changes in intensities of fluorophore-labeled IF2 and fMet-tRNAfMet to determine the effects on both 30SIC formation and 30SIC conversion to 70S initiation complexes (70SIC) of a) substituting AUG with AUU, and/or b) omitting IF3, and/or c) replacing GTP with the non-hydrolyzable analogue GDPCP. We demonstrate that the presence or absence of IF3 has at most minor effects on the rate of 30SIC formation using either AUG or AUU as the initiation codon, and conclude that the high affinity of IF2 for both 30S subunit and initiator tRNA overrides any perturbation of the codon-anticodon interaction resulting from AUU for AUG substitution. In contrast, replacement of AUG by AUU leads to a dramatic reduction in the rate of 70SIC formation from 30SIC upon addition of 50S subunits. Interpreting our results in the framework of a quantitative kinetic scheme leads to the conclusion that, within the overall process of 70SIC formation, the step most affected by substituting AUU for AUG involves the conversion of an initially labile 70S ribosome into a more stable complex. In the absence of IF3, the difference between AUG and AUU largely disappears, with each initiation codon affording rapid 70SIC formation, leading to the speculation that it is the rate of IF3 dissociation from the 70S ribosome during IC70S formation that is critical to its fidelity function.
Keywords: translation initiation, IF3, fidelity, 30S initiation complex, 70S initiation complex
INTRODUCTION
Protein synthesis in prokaryotes begins with the interaction of the initiator fMet-tRNAfMet and mRNA with the small (30S) ribosomal subunit. These two ligands bind stochastically to the 30S ribosomal subunit without initially interacting with each other, yielding a “30S pre-initiation complex”. A rate-limiting first-order conformational transition, which is kinetically controlled by all three initiation factors IF1, IF2 and IF3 (for reviews see references 1 and 2), transforms the 30S pre-initiation complex into a “30S initiation complex” (30SIC). In this transition, the canonical initiation codon (i.e., AUG or, more rarely, GUG or UUG)3 moves from an initial “standby” site to the P-decoding site4-8 where it base pairs with the 3'-UAC-5' anticodon of initiator tRNA.
The initiation factors play different roles in facilitating 30SIC9. IF3 influences both the on and off rates of fMet-tRNAfMet binding, increasing the latter in atypical 30S complexes, such as those containing a non-canonical initiation triplet (e.g., AUU or AUC). The presence of IF3 introduces a translational bias which reduces or prevents translation starting from non-canonical initiation complexes, pseudo-initiation complexes, or complexes containing leaderless mRNA1,2, preventing thereby formation of an “incorrect” 30SIC. This “fidelity function” of IF3 has also been described in vivo3,10 and provides the molecular basis for the translational auto-regulation of infC expression by IF311-13. fMet-tRNAfMet binding to the 30SIC is reversible, but becomes essentially irreversible following 30SIC association with 50S subunits and formation of the 70S initiation complex (70SIC). Conversion of reversible to irreversible fMet-tRNAfMet binding depends on release of IF3 from the ribosome13, making IF3 release a key event in 70SIC formation.
IF2 plays a complementary role to that of IF3, selectively promoting the ribosomal binding of the initiator tRNA by increasing its on-rate, while having only a marginal effect on the binding of elongator aminoacyl-tRNAs14-16. Although IF2 forms a binary complex with fMet-tRNAfMet via its C2 sub-domain (domain IV)17,18, it remains unclear whether fMet-tRNAfMet binds to 30S subunits as part of this binary complex19, or rather selectively binds to a 30S complex already containing IF220, or whether both mechanisms are possible. The combined actions of IF2 and IF3 are thus responsible for the overall fidelity of the initiation process. Although IF1 might also have a fidelity function, recent in vivo experiments21 have cast doubt on an earlier suggestion that IF1 binding within the A-site of the 30S subunit might prevent premature A-site binding of elongator aminoacyl-tRNA22.
Previous studies have, in the main, focused on the effects of initiation factors on the fidelity of 30SIC formation, and only relatively recently has attention broadened to consider the “fidelity role” that such factors might play during the transition from 30SIC to 70SIC23-27. In the accompanying article28, we present a quantitative kinetic scheme for 70SIC formation from 30SIC based on rapid kinetics measurements of five observables [light scattering, GTP hydrolysis, Pi release, and changes in the fluorescent intensities of fluorescent derivatives of IF2 (denoted IF2C) and fMet-tRNAfMet (denoted fMet-tRNAfMet(prf20)]. According to this scheme (Figure 1), the initial 70S complex formed as a result of step 1, while sufficient to support IF2-dependent GTP hydrolysis (step 2), can rather readily dissociate into 30S and 50S subunits (step 2'). The subsequent conformational change that occurs in step 3 is crucial for the two most important aspects of 70SIC formation, the stabilization of the 70S particle and the subsequent accommodation of the 3'-end of fMet-tRNAfMet within the 50S P-site (step 4). The latter is essentially the rate-determining step for 70SIC formation. The subsequent rapid and essentially irreversible loss of Pi (step 5) completes 70SIC formation.
Figure 1. A quantitative scheme for 70SIC formation, emphasizing the role of IF228.
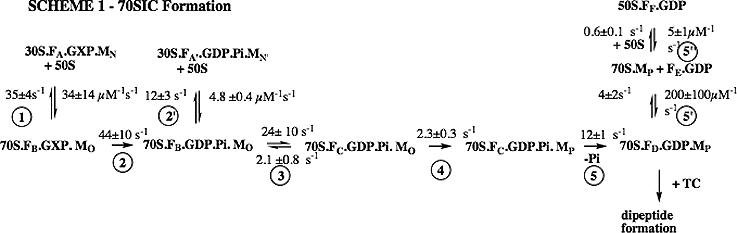
Here GXP is either GTP or GDPCP; F is IF2; M is fMet-tRNAfMet; 30S is 30S containing IF1, IF3, and mRNA; and TC is ternary complex (Phe-tRNAPhe.EF-Tu.GTP). The subscripts A-F and N-P refer to different conformations of IF2C and fMet-tRNAfMet(prf20), respectively, that have different fluorescent intensities.
In the present study we examine the effects on both 30SIC formation and 30SIC conversion to 70SIC of a) substituting the canonical AUG initiation codon with the non-canonical AUU codon, b) omitting IF3, and c) replacing GTP with the non-hydrolyzable analogue GDPCP. The effects of these changes, made singly or in combination, are determined by measurement of all or some of the same five observables used in formulating Scheme 1 (Figure 1), and the results obtained are interpreted using this scheme as a framework. We find that formation of stable 30SIC is little affected by the presence of IF3 regardless of which codon is present in the P site, but that IF3 severely retards conversion of 30SIC to 70SIC when AUU replaces AUG.
RESULTS
Rapid high affinity IF2(GTP and fMet-tRNAfMet binding to 30S complexes
In the accompanying article28 we used the fluorescence spectral change resulting from the binding of fluorescent IF2(GTP (IF2C) to 30SIC* (defined as 30S subunits containing all ligands required to form 30SIC except for IF2) to demonstrate that such binding proceeds very rapidly and with high affinity via a two-phase reaction, with an overall Kd ( = k−1k−2/k1k2) of 2.1 nM.
Substituting the non-canonical initiation triplet AUU for the canonical AUG present in the 022mRNA13 has very little effect on either the kinetics or level of IF2 binding (Figure 2A). The dependence of IF2 binding on 30SIC* concentration is similarly unaffected by the AUU for AUG substitution, with the results presented in Figure 2B permitting calculation of an overall Kd of 2.7 nM. These results are in good agreement with recent data obtained by more traditional binding studies29. The results of IF2C binding studies similar to those displayed in Figure 2B are summarized in Table 1. They show that, in the presence of fMet-tRNAfMet, the presence or absence of IF3 has only minor effects on IF2C.GTP binding to 30S subunits programmed with either AUG or AUU as the start codon, with the values determined for Kd varying over a limited range (1.9 − 5.8 nM) for all four combinations investigated. However, the affinity of IF2C.GTP for the subunit is decreased almost 20-fold in the absence of fMet-tRNAfMet and >100-fold in the absence of both IF3 and fMet-tRNAfMet (Table 1). These results, which are mainly accounted for by marked effects (up to a >30-fold increase) seen on k−1, are consistent with earlier demonstrations that both fMet-tRNA29 and IF330 increase the affinity of IF2 for the 30S subunit.
Figure 2. IF2C and fMet-tRNAfMet(prf20) binding to 30S subunits.
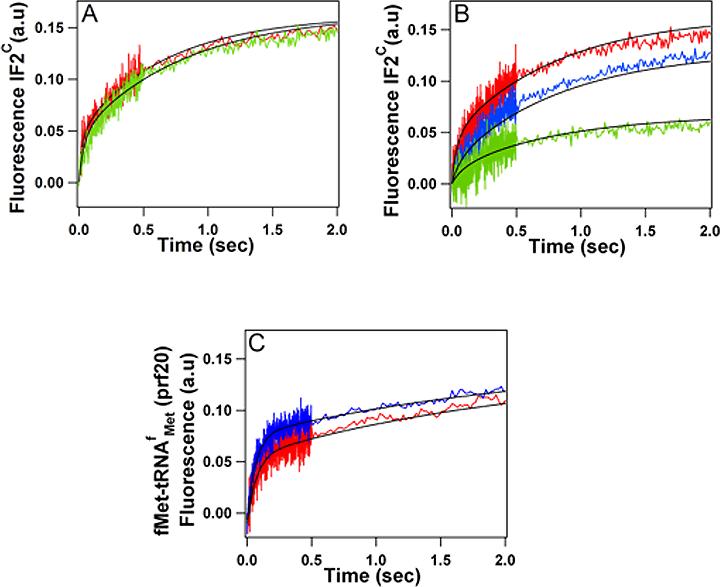
(A) IF2C·GTP binding to 30SIC* subunits programmed with AUG022mRNA (red trace) or AUU022mRNA (green trace). (B) IF2C·GTP binding to varying concentrations of 30SIC* programmed with AUU022mRNA: 0.3 μM, red trace; 0.1 μM, blue trace; 0.05 μM, green trace. (C) fMet-tRNAfMet binding to 30S subunits programmed with AUG022mRNA (red trace) or AUU022mRNA (blue trace).
Table 1.
Ligand binding to 30Sa
| Added ligand | IF3 | fMet-tRNA | mRNA | k1, μM−1s−1 | k−1, s−1 | k2, s−1 | k−2, s−1 | Kov, nM | |
|---|---|---|---|---|---|---|---|---|---|
| 1 | IF2c.GTP | + | + | AUG | 285±12 | 1.2±0.1 | 1±0.1 | 0.5±0.1 | 2.1 |
| 2 | IF2c.GTP | − | + | AUG | 310±23 | 1.2±0.24 | 0.5±0.1 | 0.75±0.10 | 5.8 |
| 3 | IF2c.GTP | + | − | AUG | 190±12 | 3.4±0.4 | 0.4±0.1 | 0.8±0.1 | 35 |
| 4 | IF2c.GTP | − | − | AUG | 111±12 | 37±5 | 1.1±0.1 | 0.9±0.1 | 273 |
| 5 | IF2c.GTP | + | + | AUU | 94±3 | 0.3±0.04 | 0.7±0.1 | 0.6±0.1 | 2.7 |
| 6 | IF2c.GTP | − | + | AUU | 79±3 | 0.1±0.03 | 0.4±0.1 | 0.6±0.12 | 1.9 |
| 7 | fMET | + | + | AUG | 14.5b | 0.45 | |||
| 8 | fMET | + | + | AUU | 15.8b | 0.43 |
All solutions contained IF1, mRNA, and 30S subunits in a molar ratio of 1.5:3:1. When present the ratio of IF3:30S was 1.5:1. GTP was 100 μM. For experiments 1 − 6, 30S concentration varied from 0.05 − 0.30 μM, IF2 was 0.15 μM. For experiments 7 and 8, concentrations were: 30S, 0.3 μM; IF2, 0.45 μM, fMet(Prf20) 0.18 μM. All concentrations are final.
in s−1
fMet-tRNAfMet(prf 20) also displays a biphasic spectral change on binding to 30S subunits to form a 30SIC complex. The rate and extent of such binding are, as in the case of IF2, virtually unaffected by substituting 022mRNA(AUU) for 022mRNA(AUG) (Figure 2C, Table 1). La Teana et al.13 also showed that fMet-tRNAfMet binds with comparable stoichiometry both to 022mRNA(AUG) and 022mRNA(AUU) programmed 30S subunits under conditions similar to those we employ, but that, in the presence of IF3, fMet-tRNA dissociates more readily on dilution of the AUU complex. With respect to 30SIC formation, it is likely that the high affinity of IF2 for both 30S subunit and initiator tRNA overrides any perturbation of the codon-anticodon interaction resulting from AUU for AUG substitution. Indeed, even in the complete absence of mRNA, fMet-tRNA bound to the 30S subunit in the presence of IF2 becomes puromycin reactive upon addition of 50S subunits23.
Rapid kinetic measures of canonical 70SIC formation
Association of a 50S subunit with the canonical 30SIC containing 022mRNA(AUG) results in the formation of the canonical 70SIC. This process is accompanied by several uni- or multi-phase changes: GTP is hydrolyzed in a single phase, Pi release and light scattering (LS) changes occur in two phases, and the fluorescence changes of both IF2C and fMet-tRNAfMet(prf20) proceed in three phases. The apparent rate constants for the changes occurring in these phases are denoted here as: GTP1 (GTPase); Pi1, Pi2 (Pi release); LS1, LS2 (light scattering); IF2−1, IF2−2, IF2−3 (IF2c fluorescence); and fMet1, fMet2, fMet3 [fMet-tRNAfMet(prf20) fluorescence].
These apparent rate constants, presented in Table2 (experiment #1), fall within three broad chronologicalphases that can be related to the specific steps described in Scheme 1 (Figure 1). The rapid phase (step 1), corresponding to the initial binding of 50S subunit to 30SIC, has a net rate constant (127 s−1) as measured by LS1, IF2−1, or fMet1. The subsequent intermediate phase includes steps 2, 2' and 3 [corresponding to GTP hydrolysis, reversible dissociation into subunits (Kd, 2.5 μM), and conformational change within 70S with IF2 fluorescence change, respectively] and is monitored by four different measures (LS2, IF2−2, fMet2, and P1) which provide similar estimates of a net rate constant (5−6 s−1). GTP hydrolysis occurs early in phase 2, and is considerably faster (GTP1 is 33 s−1) than these other events. The final, slow phase includes steps 4, 5, 5' and 5" (corresponding to conformational change of fMet within 70S, Pi release, and reversible IF2C dissociation from 70S and binding to free 50S, respectively). Its net rate constant can be measured by fMet3 and Pi2 (2 s−1), or by IF2−3 (1.2 s−1). The latter rate constant is somewhat lower since it includes changes due to steps 5' and 5"; which occur late in the slow phase and are not directly related to 70SIC formation, being important only in the presence of excess free 50S subunits.
Table 2.
Apparent rate constants in 70SIC formation
| Expt | Reaction components | Apparent rate constants (s−1) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| # | mRNA | IF3 | fMet-tRNA | GXP | LS1/IF2−1/fMet1a | GTP1 | LS2 | IF2−2 | fMet2 | Pi1 | fMet3 | Pi2 | IF2−3 |
| 1 | AUG | + | + | GTP | 127±24 | 33±3 | 6.5±1 | 5.6±0.5 | 4.9±0.5 | 4.6±0.5 | 2.2±0.2 | 1.9±0.2 | 1.2±0.1 |
| 2 | AUU | + | + | GTP | 89±9b | 32±5 | 0.36±0.04 | 4.0±0.4 | −d | 3.4±0.3 | −d | 1.1±0.1 | 1.3±0.1 |
| 3 | AUG | − | + | GTP | 108±16 | 18±2 | 6 ± 2 | 5.1±0.5 | 10 ± 3e | 5.3±0.5 | 2.9±0.3 | 2.9±0.2 | 1.6±0.2 |
| 4 | AUU | − | + | GTP | 91±9 | 12±1 | 10±1 | 9±1 | 11 ± 3 | 8±2 | 3.3±0.3 | 2.5±0.3 | 1.2±0.1 |
| 5 | AUG | + | + | GDPCP | 129±13 | nac | 5.3±0.5 | 5.2±0.5 | 4.0±0.4 | na | 1.1±0.1 | na | 1.3±0.1 |
| 6 | AUU | + | + | GDPCP | 89±10b | na | 0.47±0.05 | 3.2±0.6 | −d | na | −d | na | 1.0±0.1 |
| 7 | AUU | − | + | GDPCP | 84±8 | na | 6.3±0.6 | 7.6±0.7 | 5.9±0.8 | na | 1.7±0.2 | na | 1.2±0.1 |
In general, these three processes occur with indistinguishable apparent rate constants. Accordingly, values obtained by fitting light scattering data were used for all three accept as otherwise noted for IF2−1 (footnote b).
No rapid phase of IF2 fluorescence change was detected in these experiments,.
na, not applicable
lack of fluorescent change following initial fMet-tRNAfMet(prf20) binding prevented rate constant calculation
anomalously high value of low precision
Below we examine the effects on these apparent rate constants of imposing several changes, either singly or in tandem, on canonical 70SIC formation.
Interplay between IF3 and the initiation codon
Replacing 022mRNA (AUG) by 022mRNA(AUU) during 70SIC formation has little effect on LS1, fMet1 or GTP1, but leads to a dramatic decrease (18-fold) in LS2 (Table 2, experiment #2), as well as to a reduction in the magnitude of the initial phase of light scattering change (Figure 3B). These effects are most straightforwardly interpreted as reflecting a large reduction in the rate with which the labile 70S complex is converted into stable 70SIC via steps 3 and 4, resulting in a shift toward dissociation of 70S ribosome via the reversible reaction 2' into 30S and 50S subunits. Since the value of LS2 is considerably lower than Pi2 (Table 2), Pi release must occur from the labile 70S complex and/or from the 30S.IF2.GDP.Pi.fMet complex produced via step 2', albeit somewhat more slowly than during canonical 70SIC formation. The similarity in the values of IF2−3 and Pi2 indicates that such release is accompanied by an IF2C fluorescence change.
Figure 3. The effects of replacing AUG with AUU on measures of 70SIC formation in the presence of IF3.

(A) GTPase. (B) Light Scattering. (C) IF2C fluorescence change. (D) fMet-tRNAfMet(prf-20) fluorescence change. (E) Pi Release. AUG022mRNA (red traces), AUU022mRNA (blue traces). The gray trace in (C) is a control of 30SIC in the absence of added 50S.
Other marked differences are seen in the fluorescence changes of IF2C (Figure 3C) and fMet-tRNAfMet(prf 20) (Figure 3D). For IF2C, the rapid change upon initial 50S binding is not seen, while the 2nd and 3rd phase changes are reduced in magnitude. For fMet-tRNAfMet(prf 20), a rapid though smaller increase is still seen upon mixing 30SIC with 50S subunits (phase 1) , but the further, slower increase seen in the presence of 022mRNA (AUG) is replaced by a barely detectable fluorescence decrease in the presence of 022mRNA (AUU), even as 70SIC formation is slowly completed (Figure 3B). These effects presumably reflect alterations in the binding of fMet-tRNAfMet and IF2C to the 70S ribosome and/or to each other as a consequence of replacing cognate codon-anticodon interaction with AUG by near-cognate interaction with AUU.
When, however, IF3 is omitted, the effects caused by substituting AUU for AUG in 022mRNA are much less pronounced (Figure 4, Table 2, experiments #3 and #4). The apparent rate constants obtained with both mRNAs are similar to each other in magnitude and also similar to those observed in the formation of the canonical 70SIC, with (LS1,IF2−1,fMet1) > GTP1>(LS2, IF2−2, fMet2, P1)>(fMet3, Pi1>IF2−3). The main differences are those observed for the intermediate phase, which proceeds somewhat more rapidly in the presence of 022mRNA(AUU) (∼8.5 − 10 s−1) than in the presence of 022mRNA(AUG) (5−6 s−1). With either mRNA there is also a somewhat larger difference between fMet2, Pi2 (2.5 − 3 s−1) and IF2−3 (1.3 − 1.6 s−1) compared to what is found during canonical 70SIC formation.
Figure 4. The effects of replacing AUG with AUU on measures of 70SIC formation in the absence of IF3.

(A) GTPase. (B) Light Scattering. (C) IF2C fluorescence change (D) fMet-tRNAfMet(prf-20) fluorescence change and (E) Pi Release. AUG022mRNA (red traces), AUU022mRNA (blue traces). The gray trace in (C) is a control of 30SIC in the absence of added 50S.
There are also differences vis-à-vis canonical 70SIC formation in the fluorescence changes of IF2C (Figure 4C) and fMet-tRNAfMet(prf 20) (Figure 4D). Such differences are likely to reflect changes in the immediate environments of each fluorophore when IF3 is absent. Indeed, a cryoelectronmicroscopy reconstruction of an IF2-GDPNP-stalled 70S complex has been interpreted as indicating that IF3 is bound in a position that is very close to, and possibly in direct contact with, both fMet-tRNAfMet and IF231. In addition, at the single 50S concentration employed, the total change of light scattering in the absence of IF3 is greater for 022mRNA(AUU) than for 022mRNA(AUG) (Figure 4B), suggesting that differences of the codon-anticodon interaction may have a direct effect on the strength of 50S binding to 30SIC.
Replacement of GTP by GDPCP
Replacement of GTP by GDPCP has relatively modest effects on 70SIC formation, leading to no change in either the rate or magnitude of light scattering increases26,28 and reducing only somewhat the rate and magnitude of the fluorescence increase in fMet-tRNAfMet(prf 20) and the magnitudes of the IF2C fluorescence intensity changes during the 2nd and 3rd phases26 .
Similarly, the IF3-dependent effects seen on substitution of 022mRNA(AUG) by 022mRNA(AUU) are retained when GTP is replaced by GDPCP. Thus, substitution of AUU for AUG in the presence of GDPCP leads to large decreases both in LS2 (11-fold, Table 2, experiments #s 5, 6), and in the magnitude of the initial phase of light scattering change (Figure 5A). In addition, the effect on fMet-tRNAfMet(prf20) fluorescence change is very similar to that seen with GTP (compare Figures 5C and 3D). The effect on IF2C fluorescence change is even more dramatic than in the presence of GTP (compare Figures 5B and 3C), signaling a possible difference in the conformation of IF2.GDPCP as compared with IF2.GDP.Pi bound within the noncanonical 70S complex (see Scheme 1 in Figure 1). As is the case when GTP is present, these dramatic changes induced by substituting 022mRNA(AUU) for 022mRNA(AUG) are largely reversed when IF3 is omitted (Figure 5, Table 2, experiment #7).
Figure 5. The effects of replacing GTP with GDPCP on measures of 70SIC formation.

(A) Light Scattering. (B) IF2C fluorescence change (C) fMet-tRNAfMet(prf20) fluorescence change. Red traces AUG022mRNA + IF3; Blue traces, AUU022mRNA + IF3; Green traces, AUU022mRNA - IF3. The gray trace in (C) is a control of 30SIC in the absence of added 50S.
The interplay between IF3 and fMet-tRNAfMet
Omission of fMet-tRNAfMet weakens the binding of IF2 to 30S subunits programmed with cognate mRNA (Table 1) and prevents 70S formation (Figure 6A), with the result that the reduced GTPase activity (Figure 6B), monotonic decrease in IF2C fluorescence (Figure 6C) and very slow Pi release (Figure 6D) that result from 50S addition are due mostly to direct IF2.GTP binding to 50S subunits28. However, when both IF3 and fMet-tRNAfMet are omitted, the changes in light scattering, IF2C fluorescence, Pi release, and GTPase occurring upon 50S addition indicate that, despite much-weakened binding of IF2C to 30S subunits (Table 1), 70S formation has been at least partially restored. These results show that IF3 destabilizes the 70S structure even when the ribosomes are programmed with a canonical AUG start codon, and suggest that the associative activity of fMet-tRNAfMet counteracts such destabilization.
Figure 6. The effects of IF3 on 70S formation in the absence of fMet-tRNAfMet.
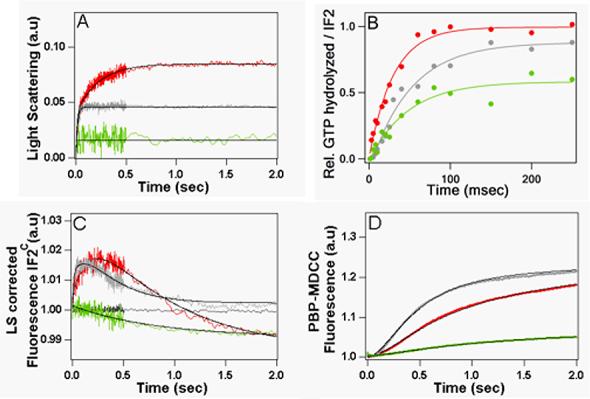
(A) GTPase. (B) Light Scattering. (C) IF2C fluorescence change and (D) Pi Release. AUG022mRNA and GTP were used in all experiments. Green traces, +IF3. Grey traces, -IF3. Red traces, reproduced from Figure 3 for comparison, are in presence of fMet-tRNAfMet and IF3.
Rates of dipeptide formation
Rates of fMetPhe formation were measured by rapidly mixing various forms of 30SIC with 50S subunits and ternary complex Phe-tRNAPhe-EF-Tu-GTP (Figure 7, Table 3). In this experiment, 70SIC formation is followed by ternary complex binding and dipeptide formation. The overall rate constant for dipeptide formation (0.18 s−1), which was determined using the canonical 30SIC complex, is virtually identical to that measured earlier24, and provides an almost direct measure of the specific rate constant for dipeptide formation, since both 70SIC formation [overall rate constant of ∼ 1.4 s−1 (Table 2)] and ternary complex binding to 70SIC32 proceed much more rapidly. When 022mRNA(AUG) is substituted by 022mRNA(AUU) the rate of peptide bond formation is decreased almost 2-fold. This relatively small decrease arises from the very large decrease in LS2 on switching to the non-canonical near cognate initiation codon (from 6.5 s−1 to 0.36 s−1, Table 2). As a result, the rates both of 70SIC formation and of the transpeptidation reaction make significant contributions to the overall rate constant for dipeptide formation. Omission of IF3 in the presence of non-canonical AUU triplet largely restores the rate constant for dipeptide formation, to 0.16 s−1, in line with the large increase in LS2. However, this rate constant is still smaller than what is seen with canonical triplet AUG in the absence of IF3 (0.24 s−1). The difference is perhaps due to a small change in the positioning of bound fMet-tRNAfMet, as reflected in the spectral changes observed in Figure 4D.
Figure 7. The effects of several substitutions on the rate of dipeptide formation from 70SIC.
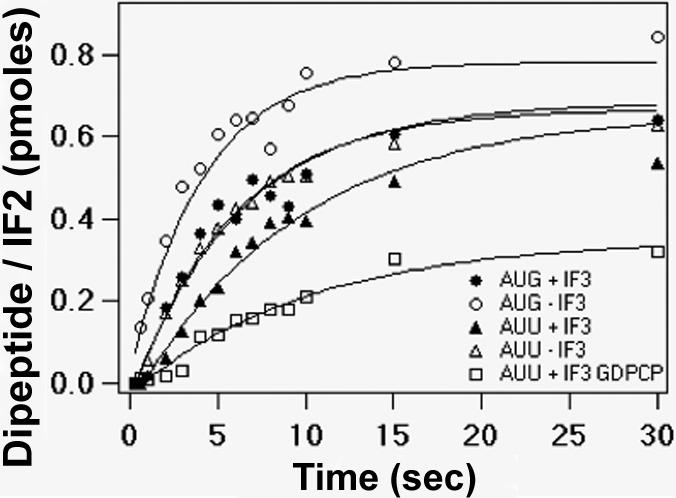
Canonical 70SIC, λ; -IF3, μ; AUU022mRNA, σ; AUU022mRNA, – IF3, Δ; AUU022mRNA, GDPCP, □.
Table 3.
Apparent rate constants for dipeptide formation
| Expt | mRNA | GXP | IF3 | kapp (s−1) |
|---|---|---|---|---|
| 1 | AUG | GTP | + | 0.18 ± 0.02 |
| 2 | AUG | GTP | − | 0.24 ± 0.04 |
| 3 | AUU | GTP | + | 0.10 ± 0.01 |
| 4 | AUU | GTP | − | 0.16 ± 0.01 |
| 5 | AUU | GDPCP | + | 0.10 ± 0.01 |
In the accompanying paper we showed that substituting GDPCP for GTP in the canonical 30SIC complex leads to no change in overall rate constant for dipeptide formation (∼0.2 s−1) but with a reduced stoichiometry28. The same pattern is seen for the noncanonical complex, in which once again the lower rate constant (0.1 s−1) vs. canonical complex reflects a large decrease in LS2 (Table 2, experiments#2 and #6).
Effect of IF3 concentration on rates of light scattering change
The results presented above provide strong evidence for the involvement of IF3 in distinguishing between canonical vs. non-canonical mRNA within the 70S initiation complex, requiring that 70S formation precede IF3 dissociation. Consistent with this view, we find the two apparent rate constants describing light scattering increase on 50S addition to 30SIC to be essentially independent of IF3 concentration over the range 0.0−3.0 μM (Figure 8), having average values similar to those shown in Table 2 (Experiment # 1). Since the second phase of light scattering change includes Scheme 1 through step 3, these results suggest that IF3 dissociation occurs during or subsequent to step 3.
Figure 8. Effect of IF3 on 70SIC formation as monitored by light scattering.

30SIC, containing increasing concentrations of IF3, was rapidly mixed with 3.0 μM 50S ribosomal subunits. IF3 concentrations were: 3.0 μM, red trace; 1.5 μM, blue trace; 0.45 μM, green trace; 0.225 μM brown trace; no IF3, grey trace. Other 30SIC components were employed at the standard final concentrations described in Materials and Methods.
DISCUSSION
As shown by our present results and in the accompanying paper28, conversion of a canonical 30SIC into a canonical 70SIC can be conveniently described as a three-phase process. In the first phase, rapid subunit association leads to formation of a labile 70S complex and induces a first conformational change within IF2, thereby activating its GTPase activity. In the second phase, the labile 70S complex either dissociates reversibly into 30S and 50S subunits or is converted to a more stable form, with the latter conversion being coupled to a second IF2 conformational change. The third phase, resulting in 70SIC formation involves movement of fMet-tRNAfMet, most plausibly to occupy the tRNA P-site within the 50S subunit, and a third IF2 conformational change largely associated with release of Pi. The second and/or third changes likely coincide with a progressive weakening of the interaction between subdomain C2 of IF2 and the acceptor end of fMet-tRNAfMet18. This flow of events is in good agreement with recent cryo-EM studies showing that IF2 bound within a 70S complex remains in strong contact with fMet-tRNAfMet prior to GTP hydrolysis31 and undergoes major conformational changes following conversion of IF2.GTP to IF2.GDP33.
During the course of 70SIC formation, IF2 is bound to 70S ribosomes in three different forms: IF2.GTP, IF2.GDP.Pi, and IF2.GDP (Figure 1). Our experiments, together with those of others, suggest that, in the presence of IF3, rapid conversion of the labile 70S complex to a more stable form has two requirements: first, that the canonical AUG initiation codon be present, rather than the noncanonical AUU initiation codon; second, that IF2 be bound as the IF2.GDP.Pi complex, or its putative structural homologue, IF2.GDPCP33. Thus, rapid conversion is achieved with ribosomes containing an AUG initiation codon and either bound IF2.GDP.Pi, formed from added IF2.GTP (Scheme 1), or bound IF2.GDPCP (Table 2, experiments #1 and #5). In contrast, such conversion is slow when AUU replaces AUG, even with bound IF2.GDPCP (Table 2, experiments #2 and #6). Further, replacement of IF2.GTP with IF2.GDP slows 70SIC formation from 30SIC by ∼ 10-fold, even with AUG as the initiation codon25.
The observed differences between AUG- and AUU-programmed ribosomes with respect to 70SIC formation depend for the most part on the presence of IF3 (Figures 3 and 4, Table 2), consistent with the well-established role of this factor in ensuring the overall fidelity of translation initiation by rejecting non-cognate or other non-standard initiation complexes1-3. Here we show that IF3 discrimination between the cognate, canonical AUG and near-cognate, non-canonical AUU initiation codons, which had been shown to occur within the 30S subunit, is also exercised during 70SIC formation. A functional role for IF3 within 70S ribosomes is supported by the results of three other studies. First, time-resolved chemical probing studies of the 30S-50S association process show that inter-subunit bridges are formed sequentially34 and that IF3 remains at least partially bound to the 30S subunit during the formation of at least some of the bridges35. Second, as already noted, there is cryo-electronmicroscopy evidence that IF3 remains ribosome-bound within an IF2-GDPNP-stalled 70S complex31. Third, monitoring of FRET signals between fluorescently-labeled IF3 and other 30S ligands, including fMet-tRNA, demonstrate that IF3 is dissociated from the 30S subunit during, rather than prior to, formation of the 70SIC (Milon, P., Konevega, A.L., Gualerzi, C.O. and Rodnina, M.V., manuscript in preparation).
A role for IF3 within 70S subunits is also consistent with the lack of effect that raising IF3 concentration has on formation of the 70S complex on addition of 50S subunits to 30SIC (Figure 8). In contrast to these results and the conclusions reached in this work, Antoun et al.27 reported that excess IF3 strongly inhibits 70S formation and concluded that IF3 dissociation must precede 70S formation. At present we can only speculate as to the reasons for this apparent discrepancy. Differences in the upstream sequences from the AUG initiation codon in the mRNA employed in both studies may be significant. The 022 mRNA used here has a relatively short Shine-Dalgarno sequence (4 nts) separated by a long spacer (9 nts) from AUG13. By contrast, the MFTI-mRNA used by Antoun et al.27 has a relatively long Shine-Dalgarno sequence (8 nts) separated by a short spacer (5 nts) from AUG. Recent results (Milon, P., Konevega, A.L., Gualerzi, C.O. and Rodnina, M.V., in preparation) demonstrate that such differences strongly impact rates of 30SIC association with 50S subunits and of initiation factor dissociation during 70SIC formation.
How does IF3 exercise its fidelity role within the 70S ribosome? In the absence of IF3, there is little codon selection between AUG and AUU (Table 2). Both initiation codons lead readily to 70SIC formation, with, somewhat surprisingly, overall reaction proceeding slightly faster with AUU than with AUG (experiments #3 and #4). This situation changes dramatically in the presence of IF3, which, while having little effect on the rate of 70SIC formation with the AUG codon (experiments #1 and #3), strongly inhibits the rate of 70SIC formation with the AUU codon (experiments #2 and #4). The similarity of the apparent 2nd phase rate constants (LS2, IF2−2, Pi1, Fmet2) for the 70S AUG complex in the presence and absence of IF3 (experiments #1 and #3) strongly implies that the conformational change taking place in this phase, corresponding to reversible step 3 in Scheme 1, precedes IF3 release. The product of step 3 is a stable 70S ribosome. As a stable 70S ribosome is sterically incompatible with bound IF336 we hypothesize that, for the 70S AUG complex, the conformational change in step 3 is followed by rapid IF3 release and the formation of additional bridges between the 30S and 50S subunits, thereby driving the reaction forward. According to this hypothesis, the inhibitory effect of IF3 on step 3 for the 70S AUU complex is due to its failure to rapidly dissociate from the ribosome as a part of step 3. This is consistent with recent results (Milon, P., Konevega, A.L., Gualerzi, C.O. and Rodnina, M.V., in preparation) showing that substituting AUU for AUG as the initiation codon retards IF3 release during 70SIC formation.
Irrespective of the precise kinetic mechanism, the question remains as to the structural basis for the IF3 fidelity function within the initially formed labile 70S ribosome. At the level of 30SIC formation IF3 could discriminate against non-canonical complexes via direct inspection of the peculiar features of the anticodon stem-loop of initiator tRNA16, 37, so direct interaction is one possibility. On the other hand, long-range, ribosome-mediated conformational change has been suggested as an explanation for IF3 discrimination against “non-standard” 30SIC 2, 35, 37 - 40. The same might be true for discrimination within a 70S ribosome. Resolution of this issue will likely require careful structural studies comparing both the 30SIC and 70SIC formed in the presence of canonical and noncanonical initiation codons.
MATERIALS AND METHODS
Materials
All materials were prepared as described previously28. 022AUU mRNA has been described12 and was prepared as previously described for 022AUG mRNA28.
Rapid kinetics measurements
All measurements were performed in 25 mM Tris-HCl pH 7.5; 70 mM NH4Cl; 30 mM KCl; 7 mM MgCl2; 1 mM DTT at 20 °C. IF2 refers to B. stearothermophilus V451C-IF2. Stopped-flow fluorescence (IF2c, fMet-tRNAfMet, Pi release) and light-scattering measurements were carried out in an SX.18MV stopped-flow spectrophotometer. Quenched flow measurements (GTPase, dipeptide formation) were carried out in a KinTec RQF-3 apparatus. All measurements were carried out as described previously28.
IF2c and fMet-tRNAfMet(prf20) binding to 30S subunits
All 30S-containing solutions were preincubated for 15 min at 37 °C prior to mixing with either IF2C or fMet-tRNAfMet(prf20). For IF2C binding experiments, IF2C was preincubated with GTP for 15 min at 37 °C, and then rapidly mixed with increasing concentrations of 30SIC*. Final concentrations of IF2C and GTP were 0.15 μM and 100 μM, respectively. Final concentrations of 30S were: 0.05 μM; 0.1 μM; 0.30 μM. 30SIC* contained, relative to 30S subunit: IF1, IF3 (when present), fMet-tRNAfMet(when present), 1.5 equivalents; 022mRNA (AUG or AUU), 3.0 equivalents. For fMet-tRNAfMet(prf20) binding experiments, final concentrations were: fMet-tRNAfMet(prf20), 0.18 μM; 30S subunits, 0.3 μM; IF1, IF2, IF3, 0.45 μM; 022mRNA (AUG or AUU), 0.9 μM; GTP 100 μM.
Measurements during 70S formation
30S complexes were preincubated for 37 °C for either 15 min (IF2C and fMet-tRNAfMet(prf20) fluorescence, light scattering, Pi release) or 10 min (GTPase) and rapidly mixed with 50S subunits. Final 50S concentration was 3.0 μM for measurements of GTPase, light scattering, and IF2C fluorescence, and 1.2 or 3 μM for measurements of Pi release and fMet-tRNAfMet(prf20) fluorescence. The standard final concentrations of 30SIC components were: 30S, 0.3 μM; IF1, IF2, IF3 (when present), fMet-tRNAfMet (when present), 0.45 μM; 022mRNA (AUG or AUU), 0.9 mM; GTP or GDPCP 100 mM. Deviations from these standard concentrations are noted below.
GTPase activity. The final concentrations of 30SIC components were 30S, 0.45 μM; IF2, 0.3 μM; [g-32P] GTP (∼1000 d.p.m./ pmol), 36 mM. GTP hydrolysis stoichiometries reported in Figures 3, 4, and 6 were normalized to the stoichiometry obtained with 70S ribosomes in the canonical system (AUG022mRNA in the presence of IF3 and fMet-tRNAfMet). This value varied from 0.7−0.9 /IF2.
Light scattering. The final concentration of IF2 was 0.15 μM.
IF2C fluorescence. IF2 was replaced by IF2C at a concentration of 0.15 μM.
fMet-tRNAfMet(prf20) fluorescence. fMet-tRNAfMet was replaced by fMet-tRNAfMet(prf20) at a final concentration of 0.18 μM.
Dipeptide formation
30S complexes were preincubated for 37 °C for 15 min and rapidly mixed with a solution containing 50S subunit and EF-Tu.Phe-tRNAPhe.GTP ternary complex. Final concentrations were: 50S subunit, 0.6 mM; ternary complex, 1.0 μM; 30S, 0.3 μM; IF1, fMet-tRNAfMet, IF3 (when present) 0.45 μM; IF2 0.15 μM; 022mRNA (AUG or AUU), 0.9 μM; GTP or GDPCP, 200 μM.
Kinetic analyses
Igor-Pro (Wavemetrics) was used to determine best fit values for GTP1 (single exponential), LS1 and LS2 (double exponential); IF-1, IF-2, and IF-3 (triple exponential), dipeptide formation (single exponential), and fMet-tRNAfMet(prf20) binding to 30S subunits (double exponential). Global fitting using the program Scientist (MicroMath Research, LC) was employed to fit values for Pi1, Pi2, fMet1, fMet2, and fMet3 and for IF2C·GTP interaction with 30SIC* via a two-step binding reaction.
ACKNOWLEDGMENTS
Supported by NIH grant GM071014 to BSC, by MIUR grants (PRIN, 2002, 2003 and 2005) to COG and by funds of the graduate program in “Biology” of the University of Camerino on behalf of C.G. and S.M. We thank Nora Zuño for excellent technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Gualerzi CO, Pon CL. Initiation of mRNA translation in prokaryotes. Biochemistry. 1990;29:5881–5889. doi: 10.1021/bi00477a001. [DOI] [PubMed] [Google Scholar]
- 2.Gualerzi CO, Brandi L, Caserta E, Garofalo C, Lammi M, La Teana A, Petrelli D, Spurio R, Tomsic J, Pon CL. Role of the initiation factors in the early events of mRNA translation in bacteria. Cold Spring Harbor Symp. Quant. Biol. 2001;66:363–376. doi: 10.1101/sqb.2001.66.363. [DOI] [PubMed] [Google Scholar]
- 3.Sussman JK, Simons EL, Simons RW. Escherichia coli translation initiation factor 3 discriminates the initiation codon in vivo. Mol. Microbiol. 1996;21:347–360. doi: 10.1046/j.1365-2958.1996.6371354.x. [DOI] [PubMed] [Google Scholar]
- 4.Canonaco M, Gualerzi CO, Pon CL. Translation initiation factors affect an alternative occupancy of a dual ribosomal binding site by mRNA. Eur. J. Biochem. 1989;182:501–506. doi: 10.1111/j.1432-1033.1989.tb14856.x. [DOI] [PubMed] [Google Scholar]
- 5.La Teana A, Gualerzi CO, Brimacombe R. From stand-by to decoding site. Adjustment of the mRNA on the 30S ribosomal subunit under the influence of the initiation factors. RNA. 1995;1:772–782. [PMC free article] [PubMed] [Google Scholar]
- 6.Yusupova G, Jenner L, Rees B, Moras D, Yusupov M. Structural basis for messenger RNA movement on the ribosome. Nature. 2006;444:391–394. doi: 10.1038/nature05281. [DOI] [PubMed] [Google Scholar]
- 7.Kaminishi T, Wilson DN, Takemoto C, Harms JM, Kawazoe M, Schluenzen F, Hanawa-Suetsugu K, Shirouzu M, Fucini P, Yokoyama S. A snapshot of the 30S ribosomal subunit capturing mRNA via the Shine-Dalgarno interaction. Structure. 2007;15:289–297. doi: 10.1016/j.str.2006.12.008. [DOI] [PubMed] [Google Scholar]
- 8.Marzi S, Myasnikov AG, Serganov A, Ehresmann C, Romby P, Yusupov M, Klaholz BP. Structured mRNAs regulate translation initiation by binding to a dedicated site on the ribosome. Cell. 2007 doi: 10.1016/j.cell.2007.07.008. in press. [DOI] [PubMed] [Google Scholar]
- 9.Wintermeyer W, Gualerzi C. Effect of E. coli initiation factors on the kinetics of NAcPhe-tRNAPhe binding to 30S ribosomal subunits. A fluorescence stopped-flow study. Biochemistry. 1983;22:690–694. doi: 10.1021/bi00272a025. [DOI] [PubMed] [Google Scholar]
- 10.Sacerdot C, Chiaruttini C, Engst K, Graffe M, Milet M, Mathy N, Dondon J, Springer M. The role of the AUU initiation codon in the negative feedback regulation of the gene for translation initiation factor IF3 in Escherichia coli. Mol. Microbiol. 1996;21:331–346. doi: 10.1046/j.1365-2958.1996.6361359.x. [DOI] [PubMed] [Google Scholar]
- 11.Brombach M, Pon CL. The unusual translational initiation codon AUU limits the expression of the infC (initiation factor IF3) gene of Escherichia coli. Mol. Gen. Genet. 1987;208:94–100. doi: 10.1007/BF00330428. [DOI] [PubMed] [Google Scholar]
- 12.Butler JS, Springer M, Grunberg-Manago M. AUU-to-AUG mutation in the initiator codon of the translation initiation factor IF3 abolishes translational autocontrol of its own gene (infC) in vivo. Proc. Natl. Acad. Sci. USA. 1987;84:4022–4025. doi: 10.1073/pnas.84.12.4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.La Teana A, Pon CL, Gualerzi CO. Translation of mRNAs with degenerate initiation triplet AUU displays high IF2 dependence and is subject to IF3 repression. Proc. Nat. Acad. Sci. USA. 1993;90:4161–4165. doi: 10.1073/pnas.90.9.4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Canonaco MA, Calogero RA, Gualerzi CO. Mechanism of translational initiation in prokaryotes. Evidence for a direct effect of IF2 on the activity of the 30S ribosomal subunit. FEBS Lett. 1986;207:198–204. doi: 10.1016/0014-5793(86)81488-0. [DOI] [PubMed] [Google Scholar]
- 15.Gualerzi CO, Wintermeyer W. Prokaryotic initiation factor 2 acts at the level of the 30S ribosomal subunit. A fluorescence stopped-flow study. FEBS Lett. 1986;202:1–6. [Google Scholar]
- 16.Hartz D, Binkley J, Hollingsworth T, Gold L. Domains of initiator tRNA and initiation codon crucial for initiator tRNA selection by Escherichia coli IF3. Genes Dev. 1990;4:1790–1800. doi: 10.1101/gad.4.10.1790. [DOI] [PubMed] [Google Scholar]
- 17.Petersen HU, Roll T, Grunberg-Manago M, Clark BF. Specific interaction of initiation factor IF2 of E. coli with formylmethionyl-tRNAfMet. Biochem. Biophys. Res. Commun. 1979;91:1068–1074. doi: 10.1016/0006-291x(79)91989-2. [DOI] [PubMed] [Google Scholar]
- 18.Guenneugues M, Caserta E, Brandi L, Spurio R, Meunier S, Pon CL, Boelens R, Gualerzi CO. Mapping the fMet-tRNA binding site of initiation factor IF2. EMBO J. 2000;19:5233–5249. doi: 10.1093/emboj/19.19.5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mayer C, Köhrer C, Kenny E, Prusko C, RajBhandary UL. Anticodon sequence mutants of Escherichia coli initiator tRNA: effects of overproduction of aminoacyl-tRNA synthetases, methionyl-tRNA formyltransferase, and initiation factor 2 on activity in initiation. Biochemistry. 2003;42:4787–4799. doi: 10.1021/bi034011r. [DOI] [PubMed] [Google Scholar]
- 20.Boelens R, Gualerzi CO. Structure and function of bacterial initiation factors. Curr Protein Pept Sci. 2002;3:107–119. doi: 10.2174/1389203023380765. [DOI] [PubMed] [Google Scholar]
- 21.Croitoru VV, Bucheli-Witshel M, Isaksson LA. In vivo involvement of mutated initiation factor IF1 in gene expression control at the translational level. FEBS Lett. 2005;579:995–1000. doi: 10.1016/j.febslet.2004.12.072. [DOI] [PubMed] [Google Scholar]
- 22.Carter AP, Clemons WM, Jr., Brodersen DE, Morgan-Warren RJ, Hartsch T, Wimberly BT, Ramakrishnan V. Crystal structure of an initiation factor bound to the 30S subunit. Science. 2001;291:498–501. doi: 10.1126/science.1057766. [DOI] [PubMed] [Google Scholar]
- 23.La Teana A, Pon CL, Gualerzi CO. Late events in translation intiation. The adjustment of fMet-tRNA in the ribosomal P-site. J. Mol. Biol. 1996;256:667–675. doi: 10.1006/jmbi.1996.0116. [DOI] [PubMed] [Google Scholar]
- 24.Tomsic J, Vitali LA, Daviter T, Savelsbergh A, Spurio R, Striebeck P, Wintermeyer W, Rodnina MV, Gualerzi CO. Late events of translation initiation in bacteria: a kinetic analysis. EMBO J. 2000;19:2127–2136. doi: 10.1093/emboj/19.9.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Antoun A, Pavlov MY, Andersson K, Tenson T, Ehrenberg M. The roles of initiation factor 2 and guanosine triphosphate in initiation of protein synthesis. EMBO J. 2003;22:5593–5601. doi: 10.1093/emboj/cdg525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Antoun A, Pavlov MY, Andersson K, Tenson T, Ehrenberg M. Ribosome formation from subunits studied by stopped-flow and Rayleigh light scattering. Biol. Proced. Online. 2004;6:35–54. doi: 10.1251/bpo71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Antoun A, Pavlov MY, Lovmar M, Ehrenberg M. How initiation factors tune the rate of initiation of protein synthesis in bacteria. EMBO J. 2006;25:2539–2550. doi: 10.1038/sj.emboj.7601140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grigoriadou C, Marzi S, Kirillov S, Gualerzi CO, Cooperman BS. Quantitative kinetic scheme for 70S translation initiation complex formation. accompanying manuscript. 2007 doi: 10.1016/j.jmb.2007.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Caserta E, Tomsic J, Spurio R, La Teana A, Pon CL, Gualerzi CO. Translation initiation factor IF2 interacts with the 30S ribosomal subunit via two separate binding sites. J. Mol. Biol. 2006;362:787–799. doi: 10.1016/j.jmb.2006.07.043. [DOI] [PubMed] [Google Scholar]
- 30.Weiel J, Hershey JW. The binding of fluorescein-labeled protein synthesis initiation factor 2 to Escherichia coli 30 S ribosomal subunits determined by fluorescence polarization. J. Biol. Chem. 1982;257:1215–1220. [PubMed] [Google Scholar]
- 31.Allen GS, Zavialov A, Gursky R, Ehrenberg M, Frank J. The cryo-EM structure of a translation initiation complex from Escherichia coli. 2005;121:703–712. doi: 10.1016/j.cell.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 32.Pape T, Wintermeyer W, Rodnina MV. Complete kinetic mechanism of elongation factor Tu-dependent binding of aminoacyl-tRNA to the A site of the E. coli ribosome. EMBO J. 1998;17:7490–7497. doi: 10.1093/emboj/17.24.7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Myasnikov AG, Marzi S, Simonetti A, Giuliodori AM, Gualerzi CO, Yusupova G, Yusupov M, Klaholz BP. Conformational transitinon of initiation factor 2 from the GTP- to GDP-state visualized on the ribosome. Nature Struct. Mol. Biol. 2005;12:1145–1149. doi: 10.1038/nsmb1012. [DOI] [PubMed] [Google Scholar]
- 34.Hennelly SP, Antoun A, Ehrenberg M, Gualerzi CO, Knight W, Lodmell JS, Hill WE. A time-resolved investigation of ribosomal subunit association. J. Mol. Biol. 2005;346:1243–1258. doi: 10.1016/j.jmb.2004.12.054. [DOI] [PubMed] [Google Scholar]
- 35.Fabbretti A, Pon CL, Hennelly SP, Hill WE, Lodmell JS, Gualerzi CO. The real time path of IF3 onto and off the ribosome. Mol. Cell. 2007;25:285–296. doi: 10.1016/j.molcel.2006.12.011. [DOI] [PubMed] [Google Scholar]
- 36.Dallas A, Noller HF. Interaction of translation initiation factor 3 with the 30S ribosomal subunit. Mol. Cell. 2001;8:855–864. doi: 10.1016/s1097-2765(01)00356-2. [DOI] [PubMed] [Google Scholar]
- 37.McCutcheon JP, Agrawal RK, Philips SM, Grassucci RA, Gerchman SE, Clemons WM, Ramakrishnan V, Frank J. Location of translational initiation factor IF3 on the small ribosomal subunit. Proc. Natl. Acad. Sci. USA. 1999;96:4301–4306. doi: 10.1073/pnas.96.8.4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petrelli D, LaTeana A, Garofalo C, Spurio R, Pon CL, Gualerzi CO. Translation initiation factor IF3: two domains, five functions, one mechanism? EMBO J. 2001;20:4560–4569. doi: 10.1093/emboj/20.16.4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lancaster L, Noller HF. Involvement of 16S rRNA nucleotides G1338 and A1339 in discrimination of initiator tRNA. Mol. Cell. 2005;20:623–632. doi: 10.1016/j.molcel.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 40.Lomakin IB, Shirokikh NE, Yusupov MM, Hellen CU, Pestova TV. The fidelity of translation initiation: reciprocal activities of eIF1, IF3 and YciH. EMBO J. 2006;25:196–210. doi: 10.1038/sj.emboj.7600904. [DOI] [PMC free article] [PubMed] [Google Scholar]


