Abstract
The dynamical coupling between proteins and their hydration water is important for the understanding of macromolecular function in a cellular context. In the case of membrane proteins, the environment is heterogeneous, composed of lipids and hydration water, and the dynamical coupling might be more complex than in the case of the extensively studied soluble proteins. Here, we examine the dynamical coupling between a biological membrane, the purple membrane (PM), and its hydration water by a combination of elastic incoherent neutron scattering, specific deuteration, and molecular dynamics simulations. Examining completely deuterated PM, hydrated in H2O, allowed the direct experimental exploration of water dynamics. The study of natural abundance PM in D2O focused on membrane dynamics. The temperature-dependence of atomic mean-square displacements shows inflections at 120 K and 260 K for the membrane and at 200 K and 260 K for the hydration water. Because transition temperatures are different for PM and hydration water, we conclude that ps–ns hydration water dynamics are not directly coupled to membrane motions on the same time scale at temperatures <260 K. Molecular-dynamics simulations of hydrated PM in the temperature range from 100 to 296 K revealed an onset of hydration-water translational diffusion at ≈200 K, but no transition in the PM at the same temperature. Our results suggest that, in contrast to soluble proteins, the dynamics of the membrane protein is not controlled by that of hydration water at temperatures <260 K. Lipid dynamics may have a stronger impact on membrane protein dynamics than hydration water.
Keywords: molecular dynamics simulations, neutron spectroscopy, dynamical transition, purple membrane, bacteriorhodopsin
Proteins are animated by a multitude of motions occurring on various length and time scales. In a current model, a protein is not characterized by a single three dimensional structure but, rather, by a large number of conformations, so-called conformational substates, that interconvert via molecular motions (1). Natively unfolded proteins reflect this conformational heterogeneity to an extreme degree. The developing concept of a free-energy landscape has put some order into the complex world of protein motions (1). The energy landscape is a high-dimensional space, in which each conformational substate is defined by the coordinates of all atoms in the protein. The energy landscape is organized in a hierarchy of levels; the top level containing only a few substates that interconvert by relatively slow large-scale movements (2). Each substate at this level contains, itself, a larger number of substates, separated by smaller barriers that are sampled by more rapid motions. At the third level, the number of substates becomes very large and the barriers very small, and motions at this level have been proposed to take place on the ps–ns time scale (2). Which of the motions on different levels are crucial for biological function and whether and how they are correlated with each other are challenging current and future issues of research in protein dynamics.
One way of exploring the complex energy landscape of proteins and its relation to function is through temperature-dependent studies (3). Particularly in the cryoregime, protein motions can be teased apart by varying the temperature. An intensively studied phenomenon in this context is the so-called dynamical transition, evidenced as a break in atomic mean-square displacements (MSDs) as a function of temperature, determined by Mössbauer (4) and neutron spectroscopies (5–7) and in Debye–Waller factors refined in x-ray crystallography (8, 9). A hydration-dependent transition occurs at a temperature between 180 and 260 K (4–7, 10–12). The increased atomic flexibility above the transition has been observed to be crucial for CO escape from myoglobin (13, 14) and for substrate and inhibitor binding in ribonuclease A (15). No correlation between the transition temperature and the onset of biological activity was observed however for certain enzymes (16, 17). A second inflection in atomic MSDs, which is hydration-independent, was observed at a temperature between 100 and 150 K (10, 18). The low temperature inflection that occurs even in dry samples was also seen in molecular dynamics (MD) simulations (19) and has been attributed to the onset of methyl group rotations (20, 21).
The hydration-dependence of the dynamical transition is one of the observations that hint at the importance of the environment on macromolecular dynamics. A solvent-viscosity dependence is yet another (22, 23). Adding cryoprotectants like sugar molecules (12, 13, 21, 24, 25), glycerol (11, 26), methanol (27), or salt (28) to the solvent shifts the dynamical-transition temperature as shown by a variety of biophysical techniques. The solvent-dependence of protein motions was early termed “slaving” (29), expressing that the solvent imposes its dynamical imprint on the protein. More recently, finer details of the dynamical picture have emerged in which protein motions are termed “solvent-slaved” and “hydration-shell coupled” if their characteristic temperature-dependence follows that of bulk solvent and hydration water, respectively (2, 30). Solvent-slaving of macromolecular motions was also described for tRNA (31).
Water in the first hydration layer (hydration or interfacial water) links the protein surface and the bulk solvent, thus playing a central role in solvent-slaving. In particular, the onset of water translational diffusion on the surface of soluble proteins has been proposed, based on MD simulations, to be the driving force behind the protein dynamical transition at temperatures between 180 and 260 K (32, 33). Time-resolved MSDs of protein hydration water deduced from neutron spectroscopic data (after subtraction of the protein contribution) on the soluble protein myoglobin revealed the onset of water translational diffusion at the protein dynamical transition (34). Based on quasi-elastic neutron scattering experiments, the change in translational mobility of hydration water in a hydrated lysozyme powder at 220 K has been interpreted as a dynamic cross-over from a “strong” (Arrhenius) below, to a “fragile” (non-Arrhenius) behavior above that temperature (35). Dielectric-spectroscopy experiments on a hydrated myoglobin powder, however, have led to the conclusion that vanishing α- (structural) relaxations and not a fragile–strong transition might be at the origin of the dynamic cross-over (36). In contrast to these findings, recent neutron scattering and diffraction experiments on hydrated stacks of purple membranes (PMs) have challenged the current view that macromolecular motions respond to dynamical changes in the hydration water (37, 38). An onset of translational mobility in water beyond the first hydration layer was indirectly observed at 200 K by monitoring the lamellar spacing of PM stacks as a function of temperature. Surprisingly, the membrane motions showed dynamical transitions at ≈150 and 260 K, yet not at 200 K.
In this study, we address the issue of solvent-slaving to PM, in the most direct approach available to us. Hydrogen dynamics on the ps–ns time scale reflects the dynamics of the groups to which they are attached and strongly dominates incoherent neutron scattering. Specific hydrogen labeling in otherwise deuterated samples can therefore be used to focus on the dynamics of different parts of a complex system. PM occurs naturally in the form of a two-dimensional crystal, consisting of 75% (wt/wt) of a single protein, bacteriorhodopsin (BR), that functions as a light-activated proton pump and 25% various lipid species (reviewed in ref. 39). Completely deuterated PM was produced and hydrated with well defined layers of H2O, thus yielding dynamical information on hydration water in elastic incoherent neutron scattering (EINS) experiments, performed on the IN16 back-scattering spectrometer at the Institut Laue Langevin (Grenoble, France). A clear change in atomic MSDs of water molecules in the first hydration layer was observed at 200 K. An identically prepared control sample of natural-abundance PM hydrated in D2O showed an inflection at 120 K related to methyl-group rotations and a dynamical transition at 260 K but no inflection at 200 K. We conclude that water and membrane motions on the ps–ns time scale are not directly coupled to each other and that the latter are thus neither solvent-slaved, nor hydration-shell coupled at temperatures <260 K. MD simulations on hydrated PM indicated that the 200 K transition is due to the onset of translational diffusion of hydration-water molecules. In PM, in contrast to soluble proteins, the onset of translational diffusion in the hydration water is not directly correlated with a dynamical transition in the membrane protein BR.
Results
EINS Experiments.
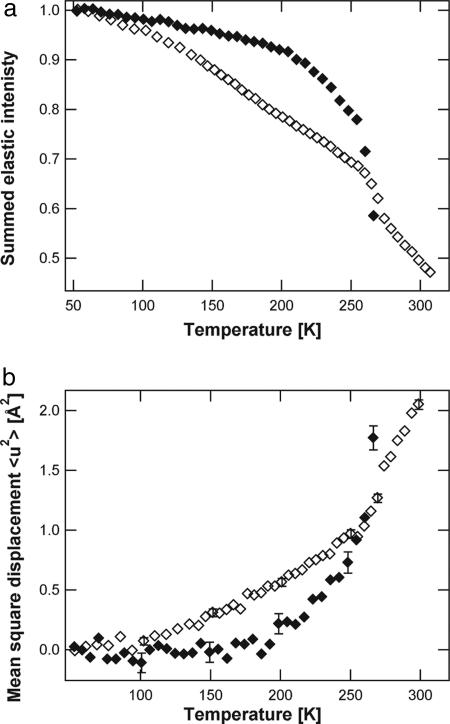
EINS experiments were carried out on PM stacks with a lamellar spacing of 62 Å at room temperature (as determined by neutron diffraction, see Materials and Methods). Given the thickness of a dry PM fragment of 49 Å, a lamellar spacing of 62 Å corresponds to an average intermembrane water layer of 13 Å. Two samples were examined: deuterated PM stacks hydrated in H2O (D-PM-H2O) and natural-abundance PM stacks hydrated in D2O (H-PM-D2O). Ninety-five percent of the total incoherent cross-section originates from water in the former, and 99% from PM in the latter [see supporting information (SI) Table 1). Consequently, water and PM dynamics can be examined separately and compared. The normalized elastic intensity summed over a defined range of Q vectors (see Materials and Methods) as a function of temperature for both D-PM-H2O and H-PM-D2O samples is given in Fig. 1a. The temperature-dependence of the summed elastic intensity is different for both samples, with inflections occurring at different temperatures. In the H-PM-D2O sample, a first inflection in the data can be seen at 120 K, whereas the temperature-dependence of the D-PM-H2O sample remains linear up to ≈200 K, indicating no significant dynamics faster than 1 ns occur below this temperature on the Å-length scale. At 200 K, a broad transition can be seen in the D-PM-H2O sample, whereas, in this temperature range, the elastic intensity from the H-PM-D2O is linear. The D-PM-H2O sample displays a sharp drop in elastic intensity between 255 and 270 K. In the same temperature region, an inflection can be seen in the summed elastic intensity of the H-PM-D2O sample.
Fig. 1.
Neutron scattering data. (a) Sum of the scattered normalized elastic intensity as a function of temperature for D-PM-H2O (filled diamonds) and H-PM-D2O (open diamonds). The sum has been performed over the ranges 0.3 < Q2 < 0.9Å−2 for D-PM-H2O and 0.3 < Q2 < 1.5Å−2 for H-PM-D2O. (b) MSDs as a function of temperature for D-PM-H2O (filled diamonds) and H-PM-D2O (open diamonds). Q-ranges are 0.3 < Q2 < 0.9Å−2 for D-PM-H2O and 0.3 < Q2 < 1.5Å−2 for H-PM-D2O.
MSDs are presented for both samples in Fig. 1b. By using the inflections in the summed elastic intensity (Fig. 1a) as a guide, the MSDs are well described by three linear regions for each sample. Below 120 K, both samples display similar MSDs. At ≈120 K, there is a kink in the MSDs of the H-PM-D2O sample, whereas those of the D-PM-H2O sample continue to increase linearly up to 200 K, where they undergo a transition to a steeper increase. Between 120 K and 260 K, the MSDs of H-PM-D2O sample are larger than those of the D-PM-H2O sample. At ≈260 K, both samples undergo a transition. Above 270 K, MSDs from the D-PM-H2O sample can no longer be calculated from the recorded data; the water molecules are now moving too far within the time scale defined by the instrument's energy resolution to be visible in the accessible space scale (40).
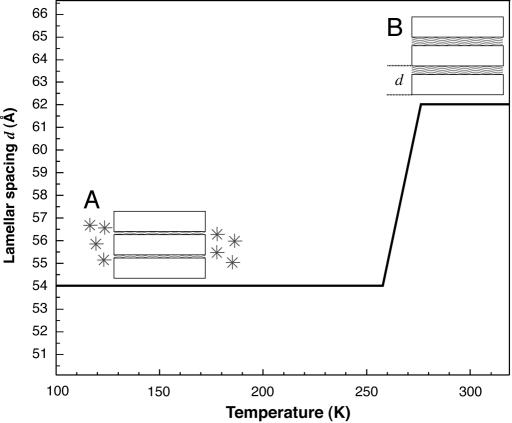
It should be noted that the lamellar spacing does not remain constant in the temperature range explored. During slow cooling from room temperature to 50 K, i.e., before the subsequent data collection during heating, the lamellar spacing of 62 Å has been shown to decrease at ≈270 K to reach a minimum value of 54 Å at 250 K (37, 41). The decrease in lamellar spacing has been identified to be due to water leaving the intermembrane space to form crystalline ice outside the regular stack of membranes. From the initial amount of intermembrane water at room temperature, ≈40% remains in contact with the membrane surfaces, and 60% has formed crystalline ice at temperatures <260 K. Upon subsequent heating, the lamellar spacing of 54 Å first remains constant and then increases between 260 K and room temperature to recover the initial value of 62 Å as the ice melts and water flows back into the membrane stack. During the data acquisition on heating in the present work, at low temperatures, scattering from the water is divided into two contributions, that of intermembrane water, and that of ice. For the D-PM-H2O sample, the corresponding contributions to the total incoherent scattering signal are ≈36% and 59%, respectively (see SI Table 1). The behavior of the lamellar spacing upon heating is presented schematically in Fig. 2.
Fig. 2.
Schematic representation of the lamellar spacing d of a hydrated PM stack as a function of temperature (after refs. 37 and 47). PM is sketched as open rectangles intercalated by hydration-water layers depicted as waved lines (A and B). The lamellar spacing d is 54 Å at 100 K (A) and 62 Å at 300 K (B), with a transition from A to B starting at ≈260 K (solid line). Part of the hydration water left the intermembrane space during slow cooling and is present as crystalline ice (depicted by asterisks in A) outside the membrane stacks at 100 K (A).
MD Simulations.
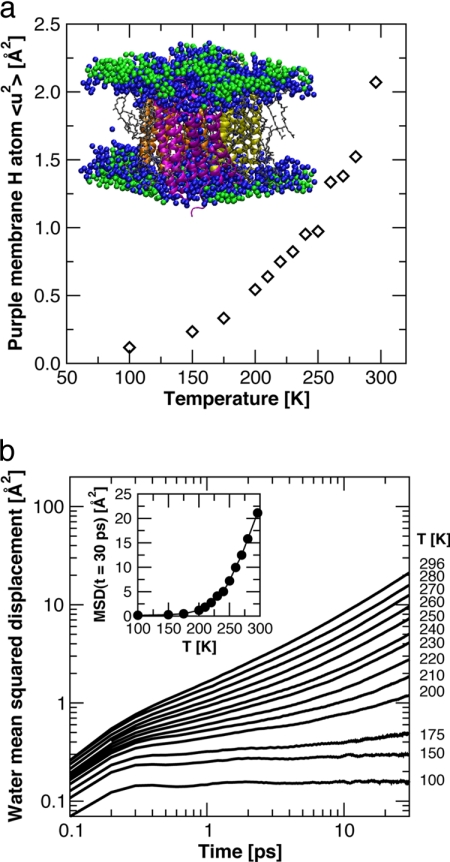
To further investigate the temperature-dependent changes in water dynamics, a series of MD simulations of an all-atom model of PM (see Materials and Methods for details) was performed over a range of temperature spanning the inflections in MSDs. A snapshot of the system simulated and a plot of the temperature dependence of MSDs, of the nonexchangeable hydrogens in PM (protein and lipid), averaged over 5 ns, are shown in Fig. 3a. Comparison with the corresponding neutron scattering data (Fig. 1b) shows that the simulations were capable of semiquantitatively reproducing the experimentally measured changes in the dynamics of PM as a function of temperature on the ns time scale.
Fig. 3.
Molecular dynamics simulation data. (a) Temperature-dependence of the mean square displacements of the nonexchangeable hydrogen atoms of the protein and lipid molecules computed from the MD simulation trajectories. Each point represents an average over 5 ns. The Inset shows a snapshot of the unit cell from one of the simulations, with the three BR monomers colored magenta, orange, and yellow, the lipid molecules gray, the water molecules in the first solvation shell (defined as within 4 Å of a heavy atom (42), taking periodic boundary conditions into consideration) of protein and/or lipid molecules blue, and the remaining water molecules green. (b) Time-evolution of the MSDs of centers-of-mass of water molecules in the first solvation shell (colored blue in a), averaged over water molecules and time origins. The Inset plots the values of the MSDs at t = 30 ps versus temperature. A dynamical transition is evident at ≈200 K.
The time evolution of the MSDs of the centers-of-mass of the water molecules in the first solvation shell (colored blue in the snapshot in Fig. 3a) of the protein and lipid molecules are plotted in Fig. 3b for each temperature. Each of the MSD curves displays a rapid initial rise, corresponding to ballistic motion, at very short times (<0.3 ps). At longer times, the MSDs exhibit qualitatively different behavior on the time scale of tens of ps, depending on whether the temperature is above or below ≈200 K. Below ≈200 K, after the initial, subpicosecond rise, the MSDs are essentially flat, and this indicates that the water molecules are in a structurally arrested, glass-like state. Above ≈200 K, the MSDs begin to curve upward after a few ps, and this indicates the onset of translational diffusion, with a diffusion rate that increases with temperature. Note that, even at room temperature, the slope (on the log–log plot) of the MSD at long time is less than unity. This is a signature of anomalous diffusion (i.e., MSD(t) ∼ tα, with α < 1) imparted by the roughness and chemical heterogeneity of the surface presented by the protein and lipid molecules (42), as well as the effects of confinement of the water molecules between the membranes in the PM stack. Similar results (data not shown) are obtained for the remainder of the water molecules in the system (colored green in Fig. 3a). The extent of translational diffusion on a given time scale begins to increase rapidly with temperature above ≈200 K. This can be seen in the Inset of Fig. 3b, where it is evident that the MSDs at a given time in the diffusive regime (e.g., at t = 30 ps) appear to display a dynamical transition at ≈200 K.
Discussion
Hydration water is a crucial component in the structural and dynamical connection of biological macromolecules to their environment. Understanding macromolecular function in a cellular context thus requires the dynamical coupling between hydration water and a macromolecule to be explored. The prevailing view is that dynamical changes in the hydration water, such as a glass transition, trigger a dynamical transition in the macromolecule. Here, we addressed the dynamical-coupling issue by monitoring hydration-water and macromolecular motions faster than 1 ns, on the Å length scale (see Materials and Methods for details) in PM separately with elastic incoherent neutron scattering as a function of (cryo-) temperature. Deuterating either PM or the hydration water put the focus on water dynamics and membrane dynamics, respectively (see SI Table 1). The temperature-dependence of MSDs shows inflections at 120 K and 260 K for the membrane and 200 K and 260 K for the hydration water. Hydration water and membrane motions thus display different temperature dependencies <260 K.
Hydration Water and Membrane MSDs as a Function of Temperature.
An inflection in the PM MSDs is observed at ≈120 K that is not seen in the hydration water (Fig. 1b). An inflection at 120 K was also reported for membrane motions in a completely dry PM (10). Our result confirms that the inflection at 120 K is hydration-independent and intrinsic to the membrane motions as supported also by MD simulations (43). The inflection at 120 K, which is observed in several protein dynamics neutron-scattering studies with similar energy and Q-resolutions (20, 34, 43, 44), has been attributed to the onset of methyl group rotations (20, 21). In the “frequency-window” model (45), however, it can be interpreted as a dynamical process entering the spectrometer window, rather than a true transition due to the onset of a new kind of motion.
In the 120 K and 260 K range, water MSDs are smaller than those of the membrane. The membrane MSDs evolve linearly, whereas the water MSDs show a transition at 200 K. Because the lamellar spacing of 54 Å remains constant in this temperature range (Fig. 2) the transition is not due to a change in the amount of interlamellar water. Up to 260 K, ≈40% and 60% of water present in the sample are in the interlamellar space and outside the stack as crystalline ice, respectively (Fig. 2 and SI Table 1). Bulk crystalline ice measured on the same neutron spectrometer has been shown to behave as a harmonic solid up to its melting point without any transition at 200 K (46). Therefore, we ascribe the inflection at 200 K exclusively to a transition in the dynamics of water in the first hydration layer in contact with the membrane surface. A transition at ≈200 K in the translational diffusion of water is clearly evident in the MD simulation results (Fig. 3b).
Evidence of a hydration water transition in PM at 200 K has also been provided by calorimetry, dielectric spectroscopy (47), and lamellar diffraction measurements (37). The diffraction measurements show that at 200 K, intermembrane water in the second hydration layers becomes mobile enough to move along distances of the order of 1 μm, the size of the average membrane patch, on the time scale of several tens of minutes (37). The transition must activate translational motions to allow the water molecules to travel such distances, because below 200 K, the diffusion does not occur. Using a microscopic model for water displacements with translational and rotational components, the authors of ref. 48 assign the MSDs extracted from the Q range below 1 Å−1 to the translational part of the motion, which also suggests the water dynamics in the first hydration layer measured here reflect translational motion. All of the evidence described, as well as the simulation presented in this work, indicate that the onset of water translational diffusion is at the origin of the transition observed in the hydration water at 200 K. The water transition is not associated with a dynamical transition in ps–ns membrane motions.
At 260 K, both the hydration water and the membrane motions display a transition, and MSDs of water motions become as large as those of membrane motions. Crystalline ice melts and returns into the intermembrane space causing a swelling of the membrane stack (Fig. 2) (37). The dynamical transition of membrane motions at 260 K, which is absent in dry membranes (10, 44), is, as postulated, directly correlated with the solvent melting. The transition seen in the water dynamics at 200 K, both in the neutron-scattering data and the MD simulations, is not sufficient to provoke a transition in the membrane.
Dynamical Coupling of Membrane and Hydration-Water Motions.
The dynamical transition in the hydration water at 200 K does not correlate with a dynamical transition in the membrane. We conclude that water and membrane motions on the ps–ns time are decoupled at temperatures up to 260 K, where a hydration-dependant dynamical transition takes place in the membrane. We note, however, that our data do not allow us to exclude the possibility that the transition in hydration water dynamics on the ps–ns time scale at 200 K triggers a transition in membrane motions on a slower or faster time scale. The onset of water translational mobility at 200 K with no transition in membrane motions at the same temperature is in apparent contradiction to previous MD simulations, where the translational component of water dynamics on the time scale of tens of ps was found to drive the protein dynamical transition (32, 33, 49). However, those simulations were performed on soluble proteins, and not on a heterogeneous system of a membrane protein and surrounding lipids. Our data indicate that the dynamical coupling to hydration water of membrane proteins is different from that of soluble proteins.
The decoupling of water and PM dynamics below 260 K is also reflected in the temperature dependence of the functional BR photocycle. The L photo-intermediate can be reached at 155 K, whereas the M intermediates, M1 and M2, are accessed at 230 K and 260 K, respectively (50). Therefore the 200 K transition does not correspond to a characteristic temperature at which one intermediate state converts to another. The dynamical transition at 260 K, however, correlates with the build-up of the key intermediate M2.
Incoherent scattering from the hydrogenated PM in D2O is dominated to ≈75% (see SI Table 1) by protein (bacteriorhodopsin) dynamics. The lipids may undergo dynamical transitions at temperatures different from 120 K and 260 K, which may be hidden by the dominant contribution of BR to the scattering signal. It should be noted that BR itself is already dynamically heterogeneous, as shown by neutron scattering on specifically labeled BR (18). The water molecules in contact with PM do not form a hydration layer with a constant thickness as suggested by the simplified scheme in Fig. 2, because protein loops and lipid head groups protrude into the hydration shell. On average, the protein extends ≈1.5 Å from the lipid level (51), and water predominantly hydrates the lipid head groups (52). A possible explanation for the decoupling between membrane and hydration-water motions below 260 K is that the lipid head groups may act as a dynamical buffer, shielding the inserted proteins from the water dynamics. Unlike soluble proteins, membrane proteins would thus be under the dynamical control of surrounding lipids rather than of hydration water. Incoherent neutron scattering on PM reconstituted with hydrogenated lipids and deuterated BR hydrated in D2O, as well as MD simulations of hydrated PM might further explore the dynamical coupling of membrane components and hydration water.
Materials and Methods
Sample Preparation and Characterization.
Two PM samples were prepared, a fully deuterated one and one natural abundance (called hydrogenated). PM samples were isolated from Halobacterium salinarum and purified by the method described previously (53). To produce fully deuterated PM, the standard medium was replaced by a deuterated algal medium (54). For neutron-scattering experiments, D2O in the deuterated PM sample (denoted D-PM-H2O) and H2O in the hydrogenated sample (denoted H-PM-D2O) were exchanged against H2O and D2O, respectively, by three successive centrifugation steps. The two concentrated membrane suspensions, containing ≈200 mg of PM each, were placed on 4 × 3 cm2 flat aluminum sample holders. Partial drying to ≈0.5 g of water per g of membrane as determined by weighing was achieved by confinement during 12 h in dessicators over silica gel. To ensure a further, more gentle drying process, the silica gel was then exchanged for a saturated KNO3 solution (in H2O for the D-PM-H2O sample and in D2O for the H-PM-D2O sample, respectively; yielding a relative humidity (r.h.) of 93%) for 7 days until the weight was stable. Subsequently, samples were allowed to equilibrate over pure water (100% r.h.) for 2 days. H2O was used for the D-PM-H2O sample and D2O for the H-PM-D2O sample. The sample holders were then sealed with indium and closed with aluminum covers to give a sample chamber of 0.3-mm thickness.
As the membrane fragments dry during sample preparation, they orient parallel to the surface of the sample holder, forming a stack of membranes with a one-dimensional periodic repeat distance, called the lamellar spacing. Both the D-PM-H2O and the H-PM-D2O samples were characterized by a lamellar spacing of 62 Å at room temperature as measured on the diffractometer D16 at the Institut Laue-Langevin. This value corresponds to an average intermembrane water layer of 13 Å, given the average thickness of a dry PM fragment of 49 Å. The hydration level corresponds approximately to 0.3 g of water per g of PM.
EINS.
Owing to their particular wavelengths and energies, thermal and cold neutrons are a unique probe for macromolecular structure and thermal dynamics (55). The incoherent cross-section of hydrogen atoms is ≈40 times larger than that of deuterium and of all other atoms present in biological macromolecules. Consequently, hydrogen atoms dominate the incoherent scattering signal of biological samples. Because hydrogen atoms are homogeneously distributed within biological macromolecules, incoherent neutron scattering probes global (space-averaged) dynamics. However, one can focus on certain components of a sample by exploiting the above-mentioned isotope effect through specific deuterium labeling. Studying fully deuterated PM in H2O thus permits a focus almost exclusively on hydration water dynamics. Ninety-five percent of the total incoherent scattering signal of D-PM-H2O is due to the contribution of intermembrane water at room temperature (see SI Table 1). Incoherent scattering of the hydrogenated control sample, on the other hand, is dominated by the membrane signal.
The time and length scale of movements assessed is determined by the energy resolution and wave vector Q accessible by the neutron spectrometer used. In the elastic incoherent neutron scattering experiments performed here, atoms that move more slowly than a certain threshold value defined by the instrument's energy resolution are considered to be immobile. In this study, all spectroscopic measurements were performed on the IN16 backscattering spectrometer at the Institut Laue-Langevin, with an energy resolution of 0.9 μeV [full-width at half-maximum of the elastic peak (56)], and a wavelength of 6.275 Å. The Q-range accessible is 0.02–1.9 Å−1, corresponding to movements on the Å scale. At the resolution used, only hydrogen movements with characteristic times faster than 1 ns are monitored. The hydrogen atoms reflect the movements of larger groups to which they are attached, such as amino acid side chains. Performing so-called “elastic” scans, that is, recording the elastic intensity as a function of temperature, can reveal dynamical changes. In particular, a sudden drop in the elastic signal is indicative of an increase in movements faster than 1 ns on the Å length scale.
The samples were placed in an “orange” cryostat at room temperature at an angle of 135° with respect to the incident beam and then cooled to 50 K in ≈2 h. Subsequently, the elastic energy was recorded on heating from 50 K to 300 K. For the H-PM-D2O sample, the heating rate applied was 0.25 K/min between 50 K and 140 K, and 0.14 K/min between 140 K and 300 K. A constant heating rate of 0.14 K/min was used for the D-PM-H2O sample. The acquisition was continuous on heating, and the data were subsequently binned into points corresponding to a temperature step of ≈5 K. The signal from the empty sample holder was subtracted, and the data were normalized to the intensity at 50 K.
From the Q-dependence of the elastic intensity (Q = 4πsinθ/λ, in which 2θ is the angle of the scattered neutron, and λ is the neutron wavelength) the MSD 〈u2〉 of the investigated movements can be calculated according to the Gaussian approximation that is valid for Q2〈u2〉 < 2:
I(Q, ω = 0) is the elastically scattered intensity, A0 is the value of the scattering at Q = 0. For the above to be valid, the quasielastic contribution to the integrated elastic intensity is considered negligible.
Here, MSDs were calculated in the ranges 0.3 < Q2 < 1.5 Å−2 for H-PM-D2O and 0.3 < Q2 < 0.9 Å−2 for D-PM-H2O. The reduced Q-range for the D-PM-H2O sample is due to a coherent peak centered at approximately Q = 1.4 Å−1 arising from the spacing of deuterated lipids at 4.5 Å (57), which broadens with increasing temperature. The detectors corresponding to this peak are excluded from the analysis.
Examining the summed elastic intensity as a guide for interpreting MSDs was first used in neutron biological dynamics studies in ref. 27. The relationship between the sum and the MSD can be given by the expansion of the exponential of Eq. 1 into a sum. Limited to the first term, the expansion gives:
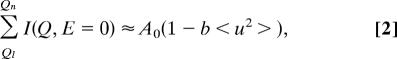 |
where b is a constant depending only on the Q values over which the sum is performed. The summed elastic intensity is therefore proportional (in a first-order approximation) to the MSD 〈u2〉. The quantity was normalized to unity at low temperature by dividing the sum by the number of detectors over which it was performed.
MD Simulations.
MD simulations of an all-atom model of PM were carried out at 13 temperatures (100 K, 150 K, 175 K, 200 K, 210 K, 220 K, 230 K, 240 K, 250 K, 260 K, 270 K, 280 K, and 296 K) spanning the experimentally observed inflections. The simulations were initiated from the PM model reported by Baudry et al. (58). A detailed description of the construction of the model was given by Baudry et al. (58), so only a brief summary is given here. The system consists of a single bacteriorhodopsin trimer, 28 lipid molecules, 41 sodium ions (for electroneutrality), and 1,924 water molecules, placed in a hexagonal unit cell, which is reproduced in three dimensions by using periodic boundary conditions. The lipids include 21 phosphatidyl glycerol phosphate molecules (PGP) and three squalene molecules arranged on the periphery of the BR trimer, one PGP molecule inside the BR trimer on the intracellular side of the membrane, and three sulfated triglycoside lipids inside the trimer on the extracellular side. All of the lipids contain the same branched, saturated acyl chains. Details of the simulations are given in SI Text.
Supplementary Material
Acknowledgments
We thank Heloisa Nunes Bordallo, Bruno Demé, and Bernhard Frick for their help during data collection at the Institut Laue-Langevin; Marek Koza for fruitful discussions; Emad Tajkhorshid for providing the coordinates for the PM model used in the MD simulations; and Jessica Morgan for performing the MD simulations. This work was supported by the Commissariat à l'Energie Atomique, the Centre National de la Recherche Scientifique, and the Université Joseph Fourier as well as an Agence Nationale de la Recherche Grant (to M.W.) (project number JC05_45685), the European Union DLAB program under contract numbers HPRI-CT-2001-50035 and RII3-CT-2003-505925, and National Science Foundation Grant CHE-0417158 (to D.J.T.). D.J.T. thanks the Université Joseph Fourier for a visiting-professorship, which facilitated his contribution to this work.
Abbreviations
- BR
bacteriorhodopsin
- EINS
elastic incoherent neutron scattering
- PM
purple membrane
- MD
molecular dynamics
- MSD
mean-square displacement.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0706566104/DC1.
References
- 1.Frauenfelder H, Sligar SG, Wolynes PG. Science. 1991;254:1598–1603. doi: 10.1126/science.1749933. [DOI] [PubMed] [Google Scholar]
- 2.Fenimore PW, Frauenfelder H, McMahon BH, Young RD. Proc Natl Acad Sci USA. 2004;101:14408–14413. doi: 10.1073/pnas.0405573101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zaccai G. Science. 2000;288:1604–1607. doi: 10.1126/science.288.5471.1604. [DOI] [PubMed] [Google Scholar]
- 4.Parak F, Knapp EW, Kucheida D. J Mol Biol. 1982;161:177–194. doi: 10.1016/0022-2836(82)90285-6. [DOI] [PubMed] [Google Scholar]
- 5.Doster W, Cusack S, Petry W. Nature. 1989;337:754–756. doi: 10.1038/337754a0. [DOI] [PubMed] [Google Scholar]
- 6.Ferrand M, Dianoux AJ, Petry W, Zaccai G. Proc Natl Acad Sci USA. 1993;90:9668–9672. doi: 10.1073/pnas.90.20.9668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fitter J. Biophys J. 1999;76:1034–1042. doi: 10.1016/S0006-3495(99)77268-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tilton RF, Jr, Dewan JC, Petsko GA. Biochemistry. 1992;31:2469–2481. doi: 10.1021/bi00124a006. [DOI] [PubMed] [Google Scholar]
- 9.Teeter MM, Yamano A, Stec B, Mohanty U. Proc Natl Acad Sci USA. 2001;98:11242–11247. doi: 10.1073/pnas.201404398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lehnert U, Reat V, Weik M, Zaccai G, Pfister C. Biophys J. 1998;75:1945–1952. doi: 10.1016/S0006-3495(98)77635-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsai AM, Neumann DA, Bell LN. Biophys J. 2000;79:2728–2732. doi: 10.1016/S0006-3495(00)76511-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paciaroni A, Cinelli S, Onori G. Biophys J. 2002;83:1157–1164. doi: 10.1016/S0006-3495(02)75239-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lichtenegger H, Doster W, Kleinert T, Birk A, Sepiol B, Vogl G. Biophys J. 1999;76:414–422. doi: 10.1016/S0006-3495(99)77208-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ostermann A, Waschipky R, Parak FG, Nienhaus GU. Nature. 2000;404:205–208. doi: 10.1038/35004622. [DOI] [PubMed] [Google Scholar]
- 15.Rasmussen BF, Stock AM, Ringe D, Petsko GA. Nature. 1992;357:423–424. doi: 10.1038/357423a0. [DOI] [PubMed] [Google Scholar]
- 16.Daniel RM, Smith JC, Ferrand M, Hery S, Dunn R, Finney JL. Biophys J. 1998;75:2504–2507. doi: 10.1016/S0006-3495(98)77694-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bragger JM, Dunn RV, Daniel RM. Biochim Biophys Acta. 2000;1480:278–282. doi: 10.1016/s0167-4838(00)00081-9. [DOI] [PubMed] [Google Scholar]
- 18.Reat V, Patzelt H, Ferrand M, Pfister C, Oesterhelt D, Zaccai G. Proc Natl Acad Sci USA. 1998;95:4970–4975. doi: 10.1073/pnas.95.9.4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hayward JA, Smith JC. Biophys J. 2002;82:1216–1225. doi: 10.1016/S0006-3495(02)75478-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roh JH, Novikov VN, Gregory RB, Curtis JE, Chowdhuri Z, Sokolov AP. Phys Rev Lett. 2005;95:038101. doi: 10.1103/PhysRevLett.95.038101. [DOI] [PubMed] [Google Scholar]
- 21.Cornicchi E, Marconi M, Onori G, Paciaroni A. Biophys J. 2006;91:289–297. doi: 10.1529/biophysj.106.081752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cornicchi E, Onori G, Paciaroni A. Phys Rev Lett. 2005;95:158104. doi: 10.1103/PhysRevLett.95.158104. [DOI] [PubMed] [Google Scholar]
- 23.Beece D, Eisenstein L, Frauenfelder H, Good D, Marden MC, Reinisch L, Reynolds AH, Sorensen LB, Yue KT. Biochemistry. 1980;19:5147–5157. doi: 10.1021/bi00564a001. [DOI] [PubMed] [Google Scholar]
- 24.Giuffrida S, Cottone G, Cordone L. Biophys J. 2006;91:968–980. doi: 10.1529/biophysj.106.081927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cordone L, Ferrand M, Vitrano E, Zaccai G. Biophys J. 1999;76:1043–1047. doi: 10.1016/S0006-3495(99)77269-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cicerone MT, Soles CL. Biophys J. 2004;86:3836–3845. doi: 10.1529/biophysj.103.035519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reat V, Dunn R, Ferrand M, Finney JL, Daniel RM, Smith JC. Proc Natl Acad Sci USA. 2000;97:9961–9966. doi: 10.1073/pnas.97.18.9961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gabel F, Weik M, Doctor BP, Saxena A, Fournier D, Brochier L, Renault F, Masson P, Silman I, Zaccai G. Biophys J. 2004;86:3152–3165. doi: 10.1016/S0006-3495(04)74363-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iben IE, Braunstein D, Doster W, Frauenfelder H, Hong MK, Johnson JB, Luck S, Ormos P, Schulte A, Steinbach PJ, et al. Phys Rev Lett. 1989;62:1916–1919. doi: 10.1103/PhysRevLett.62.1916. [DOI] [PubMed] [Google Scholar]
- 30.Fenimore PW, Frauenfelder H, McMahon BH, Parak FG. Proc Natl Acad Sci USA. 2002;99:16047–16051. doi: 10.1073/pnas.212637899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caliskan G, Briber RM, Thirumalai D, Garcia-Sakai V, Woodson SA, Sokolov AP. J Am Chem Soc. 2006;128:32–33. doi: 10.1021/ja056444i. [DOI] [PubMed] [Google Scholar]
- 32.Tournier AL, Xu J, Smith JC. Biophys J. 2003;85:1871–1875. doi: 10.1016/S0006-3495(03)74614-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tarek M, Tobias DJ. Phys Rev Lett. 2002;88:138101. doi: 10.1103/PhysRevLett.88.138101. [DOI] [PubMed] [Google Scholar]
- 34.Doster W, Settles M. Biochim Biophys Acta. 2005;1749:173–186. doi: 10.1016/j.bbapap.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 35.Chen SH, Liu L, Fratini E, Baglioni P, Faraone A, Mamontov E. Proc Natl Acad Sci USA. 2006;103:9012–9016. doi: 10.1073/pnas.0602474103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Swenson J, Jansson H, Bergman R. Phys Rev Lett. 2006;96:247802. doi: 10.1103/PhysRevLett.96.247802. [DOI] [PubMed] [Google Scholar]
- 37.Weik M, Lehnert U, Zaccai G. Biophys J. 2005;89:3639–3646. doi: 10.1529/biophysj.104.055749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weik M. Eur Phys J. 2003;12:E153–E158. [Google Scholar]
- 39.Neutze R, Pebay-Peyroula E, Edman K, Royant A, Navarro J, Landau EM. Biochim Biophys Acta. 2002;1565:144–167. doi: 10.1016/s0005-2736(02)00566-7. [DOI] [PubMed] [Google Scholar]
- 40.Gabel F. Eur Biophys J. 2005;34:1–12. doi: 10.1007/s00249-004-0433-0. [DOI] [PubMed] [Google Scholar]
- 41.Lechner RE, Fitter J, Dencher NA, Hauss T. J Mol Biol. 1998;277:593–603. doi: 10.1006/jmbi.1997.1597. [DOI] [PubMed] [Google Scholar]
- 42.Tobias DJ, Kuo IW, Razmara A, Tarek M. In: Water in Confining Geometries. Devlin JP, Buch V, editors. Berlin: Springer; 2003. pp. 213–225. [Google Scholar]
- 43.Roh JH, Curtis JE, Azzam S, Novikov VN, Peral I, Chowdhuri Z, Gregory RB, Sokolov AP. Biophys J. 2006;91:2573–2588. doi: 10.1529/biophysj.106.082214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reat V, Zaccai G, Ferrand M, Pfister C. In: Biological Macromolecular Dynamics. Cusack S, Buttner H, Ferrand M, Langan P, Timmins P, editors. Schenectady, NY: Adenine; 1997. pp. 117–121. [Google Scholar]
- 45.Becker T, Hayward JA, Finney JL, Daniel RM, Smith JC. Biophys J. 2004;87:1436–1444. doi: 10.1529/biophysj.104.042226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koza MM, Geil B, Schober H, Natali F. Phys Chem Chem Phys. 2005;7:1423–1431. doi: 10.1039/b414382e. [DOI] [PubMed] [Google Scholar]
- 47.Berntsen P, Bergman R, Jansson H, Weik M, Swenson J. Biophys J. 2005;89:3120–3128. doi: 10.1529/biophysj.104.057208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zanotti JM, Bellissent-Funel MC, Chen SH. Europhys Lett. 2005;71:91–97. [Google Scholar]
- 49.Tarek M, Tobias DJ. Biophys J. 2000;79:3244–3257. doi: 10.1016/S0006-3495(00)76557-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dencher NA, Sass HJ, Buldt G. Biochim Biophys Acta. 2000;1460:192–203. doi: 10.1016/s0005-2728(00)00139-0. [DOI] [PubMed] [Google Scholar]
- 51.Zaccai G, Gilmore DJ. J Mol Biol. 1979;132:181–191. doi: 10.1016/0022-2836(79)90390-5. [DOI] [PubMed] [Google Scholar]
- 52.Zaccai G. J Mol Biol. 1987;194:569–572. doi: 10.1016/0022-2836(87)90683-8. [DOI] [PubMed] [Google Scholar]
- 53.Oesterhelt D, Stoeckenius W. Methods Enzymol. 1974;31:667–678. doi: 10.1016/0076-6879(74)31072-5. [DOI] [PubMed] [Google Scholar]
- 54.Patzelt H, Ulrich AS, Egbringhoff H, Düx P, Ashurst J, Simon B, Oschkinat H, Oesterhelt D. J Biomol NMR. 1997;10:95–106. [Google Scholar]
- 55.Gabel F, Bicout D, Lehnert U, Tehei M, Weik M, Zaccai G. Q Rev Biophys. 2002;35:327–367. doi: 10.1017/s0033583502003840. [DOI] [PubMed] [Google Scholar]
- 56.Frick B, Gonzalez M. Physica B Condensed Matter. 2001;301:8–19. [Google Scholar]
- 57.Henderson R. J Mol Biol. 1975;93:123–138. doi: 10.1016/0022-2836(75)90123-0. [DOI] [PubMed] [Google Scholar]
- 58.Baudry J, Tajkhorshid E, Molnar F, Phillips J, Schulten K. J Phys Chem B. 2001;105:905–918. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.