Abstract
Circadian clocks are composed of central oscillators, input pathways that transduce external information to the oscillators, and output pathways that allow the oscillators to temporally regulate cellular processes. Little is known about the output pathways. In this study, we show that the Neurospora crassa osmosensing MAPK pathway, essential for osmotic stress responses, is a circadian output pathway that regulates daily rhythms in the expression of downstream genes. Rhythmic activation of the highly conserved stress-activated p38-type MAPK [Osmotically Sensitive-2 (OS-2)] by the N. crassa circadian clock allows anticipation and preparation for hyperosmotic stress and desiccation that begin at sunrise. These results suggest a conserved role for MAPK pathways in circadian rhythmicity.
Keywords: circadian output pathway, circadian rhythm, Neurospora crassa, osmotic stress phosphorelay
Circadian clocks, which contain input pathways, oscillators, and output pathways, are evolutionarily widespread and provide an adaptive advantage to organisms by permitting the anticipation of, and preparation for, predictable daily rhythms in light and temperature that occur as a result of Earth's rotation (1–3). Circadian clocks regulate daily rhythms in processes ranging from gene expression and enzyme and hormone production to sleep/wake cycles (4, 5).
The circadian clock system of Neurospora crassa is one of the best-understood circadian models (6–9). The N. crassa FREQUENCY/WHITE COLLAR complex (FRQ/WCC) oscillator is comprised of an autoregulatory transcriptional/translational feedback loop involving the frequency (frq) and white collar (wc-1, wc-2) genes and their protein products. A key feature of this oscillator is that it establishes rhythms in protein accumulation of FRQ. The rhythms in FRQ protein levels, augmented by posttranslational modifications, are necessary for most circadian rhythmicity in the organism.
Time-of-day information is passed from oscillators through output pathways to control rhythmic expression of clock-controlled genes (ccgs; ref. 10). To date, >180 ccgs have been identified in N. crassa (10–14), including genes associated with rhythms in asexual spore development (conidiation; ref. 15), metabolism (16), pheromone production (17), and stress responses (18). However, only a handful of these ccgs have been studied in depth, and few details of the output pathways from N. crassa or any other organism's circadian oscillator are known.
We previously carried out a genetic selection to obtain mutations that lie in the output pathway(s) from the N. crassa FRQ/WCC oscillator and that alter expression levels of the morning-specific ccg-1 gene (19). Three of the mutant strains isolated [circadian output pathway (COP)1-2, 1-3, and 1-4] were of particular interest because, in addition to displaying constitutively low and arrhythmic levels of ccg-1 mRNA, they displayed a period defect in the developmental rhythm and a cell lysis phenotype on agar slants. These additional phenotypes suggested that the genes mutated in these strains function in a pathway(s) that regulates the expression of several ccgs, not just ccg-1, because a ccg-1-null strain displays no discernable phenotypes (20). In this work, we show that (i) response regulator-1 (rrg-1), encoding a response regulator (RR), is the gene mutated in the COP1–4 mutant strain; (ii) the N. crassa osmotically sensitive (OS) MAPK pathway, required for surviving conditions of high osmolarity, and in which rrg-1 functions, is regulated by and acts as an output pathway from, the circadian clock, and (iii) the clock is not required to mount an acute response to hyperosmotic conditions. Circadian regulation of this pathway may allow the organism to anticipate and prepare for daily fluctuations in environmental osmolarity.
Results
The COP1–4 Phenotype Is Due to a Mutation in rrg-1.
Genetic mapping of the mutation in the COP1–4 strain placed the mutation on linkage group 1 between the mat locus and arg-1 [supporting information (SI) Fig. 7]. Examination of this region of the physical map (www.bioinf.leeds.ac.uk/∼gen6ar/newgenelist/genes/gene_list.htm) revealed a single locus, rrg-1, with mutant phenotypes similar to those of the COP1–4 strain (19, 21).
The rrg-1 ORF is predicted to encode a 1,114-aa protein with a C-terminal receiver domain typical of those in phosphoryl-accepting RR proteins (21). RRG-1 is part of an OS pathway in N. crassa that is homologous to the Saccharomyces cerevisiae high-osmolarity glycerol (HOG) cascade and analogous to the p38 MAPK pathway in mammalian cells (22, 23). RRG-1 functions downstream of the hybrid histidine kinase OS-1 (and possibly other histidine kinases) in N. crassa (21, 24). The current model is that OS-1 undergoes autophosphorylation in response to osmotic shock. The phosphoryl group is transferred to a receiver domain on the hybrid kinase and then transferred to the histidine phosphotransferase, which shuttles the phosphoryl group to RRG-1 (21). RRG-1 modulates the activity of a downstream MAPK cascade that includes OS-4 (MAPKKK), OS-5 (MAPKK), and OS-2 (MAPK) (25, 26). The MAPK cascade regulates downstream target genes that encode components needed to survive conditions of high osmolarity, as well as for conidial integrity, sexual development, and fungicide sensitivity (21).
Sequencing of the rrg-1 locus in the COP1–4 mutant strain revealed a C to T point mutation at nucleotide 2,710 of the predicted ORF resulting in a premature stop at codon 904 (SI Fig. 7). This mutation would result in an RRG-1 protein that is truncated before the conserved aspartyl residue necessary for phosphotransfer in other RR proteins (21). An rrg-1 knockout strain (Δrrg-1), created by replacement of the rrg-1 locus with the bacterial hygromycin-resistance gene (hph; ref. 21), was crossed to the wild-type clock strain bd (hereafter referred to as “wild type”) to obtain the bd Δrrg-1 strain (referred to throughout as “Δrrg-1; SI Table 1).
On minimal agar slants, both the COP1–4 strain and the Δrrg-1 strain displayed a cell lysis phenotype in which the conidiospores eventually turned deep orange due to leakage of cytoplasmic contents, including the carotenoid pigments (Fig. 1A; ref. 21). Additionally, on growth tubes (race tubes) used to measure the conidiation rhythm, both the COP1–4 and Δrrg-1 strains showed a 1-h period shortening as compared with wild-type strains (Fig. 1B) and a delay in conidiation upon transfer to constant darkness (DD). The wild-type strain formed conidiospores on the first day in DD and each day thereafter. The COP1–4 strain conidiated very little, if at all, on the first day in DD but conidiated normally each day thereafter. The Δrrg-1 strain did not form normal conidial bands until the third day in DD.
Fig. 1.
The COP1–4 strain contains a mutation in the rrg-1 locus. (A) Phenotypes of the COP1–4 and Δrrg-1 strain and these strains transformed with plasmid pCJ2 (COP1–4 + rrg-1 and Δrrg-1 + rrg-1), on slants grown in DD at 34°C for 2 days. The darker orange-red pigmentation in COP1–4 and Δrrg-1 strains is due to leakage of carotenoids from ruptured conidiospores. Note that in the Δrrg-1 strain, many of the conidiospores have fallen to the bottom of the tube. (B) Both the COP1–4 strain and the Δrrg-1 strain display a 1-h period defect on race tubes. The yellow and black bar indicates time in constant light (LL) and DD, respectively. Each strain was inoculated and grown in LL for 24 h before transfer to DD at 25°C, after which time the growth front was marked every 24 h (black lines). To more clearly visualize the period differences, the second- and third-to-last growth front markings have been erased. The period of each strain in hours ± SEM is given. The difference between wild type and COP1–4 or Δrrg-1 is significant, with P < 0.01; the differences between wild-type and COP1–4 + rrg-1 or Δrrg-1 + rrg-1 are not significant (one-way ANOVA, Dunnett's test). (C) The COP1–4 and Δrrg-1 strains are osmotically sensitive. Strains were inoculated onto VM with or without the indicated osmolyte and grown in DD at 34°C for 2 days. Strains are identified on the top, and media conditions for each strain are indicated on the wild-type strain plate.
The Δrrg-1 strain is sensitive to high osmolarity (21). We therefore tested whether the COP1–4 mutant strain is also sensitive to hyperosmotic conditions. The wild-type strain grew on all media examined, whereas only media without an osmolyte supplement supported growth of the COP1–4 and Δrrg-1 mutant strains (Fig. 1C).
Each of the above phenotypes was rescued in either the COP1–4 mutant strain or the Δrrg-1 strain after transformation with plasmid pCJ2, which contains the wild-type rrg-1 locus (Fig. 1; ref. 21). Together, these data indicated that rrg-1 is the locus mutated in the COP1–4 strain.
RRG-1 Functions in an Output Pathway to Regulate ccg-1 Rhythmicity.
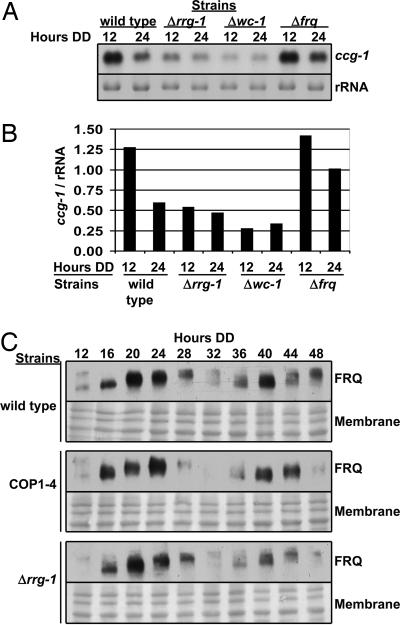
In wild-type strains, ccg-1 mRNA accumulates with a circadian rhythm, with peak levels occurring ≈12 h after entering DD (DD12) and a trough in ccg-1 mRNA levels occurring around DD24 (10). In the COP1–4 strain, ccg-1 mRNA levels are arrhythmic and constitutively low (19). Similar to COP1–4, ccg-1 mRNA levels were arrhythmic and constitutively low at all times of day in the Δrrg-1 strain (Fig. 2A and SI Fig. 8). Furthermore, in a Δfrq strain, ccg-1 mRNA levels are arrhythmic and high (although variable; Fig. 2A, SI Fig. 8, and ref. 19), whereas in a Δwc-1 strain, the levels of ccg-1 mRNA remained low at all times of day (Fig. 2A and SI Fig. 8). The low levels of ccg-1 mRNA observed in the Δwc-1 strain appear to contradict our previous report that the levels of ccg-1 mRNA are constitutively high in a WC-1 mutant strain (19); however, this difference is likely due to the fact that the WC-1 mutant strain used in the 2004 study produces a truncated protein that may have partial activity (27). These data indicated that RRG-1 is required for clock regulation of ccg-1 and suggested a positive role for WC-1 and a negative role for FRQ in regulating ccg-1 mRNA accumulation.
Fig. 2.
The FRQ/WCC oscillator is functional but unable to drive rhythmic accumulation of ccg-1 mRNA in an rrg-1 mutant strain. (A) Northern blot showing levels of ccg-1 mRNA at DD12 and DD24 in the indicated strains. The ethidium bromide-stained gel (rRNA) is shown as a loading control. (B) Bar graph of ccg-1 mRNA levels normalized to rRNA from the experiment shown in A. The hours in DD are on the x axis, and the strains are indicated below. (C) Western blot of FRQ protein in the indicated strains (on the left). Hours in DD are shown above the blots. Amido black-stained membranes are shown as loading controls. These experiments were repeated three times with identical results.
Similar to ccg-1, the ccg-9 gene (encoding trehalose synthase) and the ccg-2 gene (encoding a hydrophobin that forms a hydrophobic layer on the surface of conidia) are regulated by the clock and by acute osmotic stress (refs. 10, 19, and 28; SI Fig. 8). In a Δrrg-1 strain, ccg-9 levels were arrhythmic and low (SI Fig. 8). Previous experiments that examined ccg-2 mRNA levels in the COP1–4 mutant strain revealed arrhythmic expression of ccg-2 mRNA (19). In the Δrrg-1 strain, ccg-2 mRNA was arrhythmic in most (four of five) experiments (data not shown). However, in one experiment, a low-amplitude (2×) rhythm in ccg-2 mRNA accumulation was observed, suggesting that circadian regulation of ccg-2 is more complex and may involve multiple output pathways from the clock. Together, these data support the conclusion that the OS pathway is an output of the clock and regulates rhythmicity of a subset of ccgs.
If rrg-1 is part of an output pathway from the clock, the absence of rrg-1 should not alter the function of the FRQ/WCC oscillator. To test this, we examined FRQ protein accumulation in DD. The accumulation and phosphorylation of FRQ protein were rhythmic in the COP1–4 and Δrrg-1 strains, and this pattern was essentially indistinguishable from that of wild-type strains (Fig. 2B; ref. 29). These data indicated that the output from the oscillator, but not the central oscillator itself, is impaired in the Δrrg-1 strain.
The N. crassa OS Pathway Is Regulated by the Circadian Clock.
In the OS pathway, RRG-1 regulates phosphorylation of the OS-2 MAPK through the MAPK cascade, and phosphorylation of OS-2 is required for activity of the pathway. By using antibodies that differentially recognize only phosphorylated OS-2 protein (phospho-OS-2) or both phosphorylated and unphosphorylated OS-2 protein (total OS-2, ref. 21), we examined the accumulation and phosphorylation state of this protein over the course of 2 days in DD. The levels of OS-2 protein remained fairly constant in both the wild-type and Δrrg-1 strains (Fig. 3A). However, a robust circadian rhythm in the levels of phospho-OS-2 in the wild-type strain was observed (Fig. 3B). Phospho-OS-2 remained below detectable levels at all times of day in Δrrg-1 (Fig. 3B) and COP1–4 mutant strains (data not shown).
Fig. 3.
Phosphorylation, but not accumulation, of OS-2 protein displays a circadian rhythm. (A) Western blots showing accumulation of total (both phosphorylated and unphosphorylated) OS-2 protein. (B) Western blots showing accumulation of phosphorylated OS-2 protein. In both, hours in DD are indicated at the top, and the strains are indicated at the left. Amido black-stained membranes are shown as loading controls. The experiments shown were carried out simultaneously for comparison of levels and phosphorylation of OS-2 and were repeated three times with similar results. The lower level of total OS-2 protein in the Δrrg-1 strain at DD16 in A was not reproducible.
To determine whether the FRQ/WCC oscillator is required for rhythmic phosphorylation of OS-2, we examined both total and phospho-OS-2 levels in strains lacking FRQ or WC-1. Levels of OS-2 protein were comparable in wild-type and Δfrq strains; however, there was a reduction in total OS-2 protein in the Δwc-1 strain (Fig. 4A), and phospho-OS-2 was low or undetectable at all times of day (Fig. 4B). Phospho-OS-2 was present at all times of day in the Δfrq strain, with levels comparable to the peak in the wild-type strain (Fig. 4B).
Fig. 4.
Rhythmic accumulation of phospho-OS-2 depends on the FRQ/WCC oscillator. Western blots showing amounts of total (A) or phosphorylated (B) OS-2 protein in the indicated strains (on the left). Hours in DD are shown at the top. Amido black-stained membranes are shown for loading controls. The experiments shown were carried out simultaneously for comparison of levels and phosphorylation of OS-2 between strains and were repeated three times with similar results. The lower level of phospho-OS-2 in the Δfrq strain at DD20 in B was due to a bubble artifact. In addition, the levels of phospho-OS-2 at DD8 in the Δ wc-1 strain in B were not observed in all experiments.
In the wild-type strain, levels of phospho-OS-2 were rhythmic with peak amounts occurring at about the same time of day as the peak in ccg-1 mRNA levels (DD12; Figs. 2A and 4B). Strains with constitutively low (or no) phosphorylated OS-2 (Δrrg-1 and Δwc-1) also displayed constitutively low levels of ccg-1 mRNA (Figs. 2A, 3B, and 4B). Conversely, the Δfrq strain, which displayed constitutively elevated (arrhythmic) levels of phospho-OS-2, also displayed constitutively elevated levels of ccg-1 mRNA (Figs. 2A and 4B). The correlation between the levels of phospho-OS-2 and ccg-1 mRNA observed in these strains indicated that the OS pathway functions as an output pathway that connects the FRQ/WCC oscillator to the rhythmic expression of ccg-1.
FRQ/WCC Oscillator Is Not Necessary for an Osmotic Stress Response.
To determine whether a response to osmotic shock depends on a functional FRQ/WCC oscillator, we examined phospho-OS-2 and ccg-1 mRNA levels in wild-type and clock mutant strains. Phospho-OS-2 and ccg-1 mRNA levels, but not total OS-2 protein levels, are induced in response to treatment with 4% NaCl in wild-type strains (Fig. 5; refs. 20 and 21). Phospho-OS-2 is detectable within 5 min, and ccg-1 mRNA accumulates significantly within 1 h after 4% NaCl treatment. The induction of phospho-OS-2 and ccg-1 mRNA by 4% NaCl depends on rrg-1 (Fig. 5 B and C; ref. 21).
Fig. 5.
rrg-1, but not frq or wc-1, is necessary for osmotic induction of phospho-OS-2 and ccg-1 mRNA levels. Western blots showing total (A) or phospho-OS-2 (B) in response to hyperosmotic conditions. Time 0 = DD 24. Strains (on the left) and time in the presence of 4% NaCl (Top) are shown. The asterisks indicate treatment with sterile distilled water instead of 4% NaCl as a control. Amido black-stained membranes are shown as loading controls. The Western blots in A and B were carried out simultaneously for comparison of levels and phosphorylation of OS-2 between strains. Note that the levels of total OS-2 and phospho-OS-2 are lower in Δwc-1 and higher in Δfrq as compared with the wild-type strain. (C) Northern blot of ccg-1 mRNA levels in response to hyperosmotic conditions. Strains and time in 4% NaCl are given across the top, and two film exposures of 1.5 and 7.5 h (long exposure) are shown. The ethidium bromide-stained gel (rRNA) is shown as a loading control. These experiments were repeated three times with similar results.
Despite differences in initial levels of phospho-OS-2 or ccg-1 mRNA (time 0; very low in Δwc-1 and high in Δfrq as compared with the wild-type strain), 4% NaCl treatment resulted in increased phospho-OS-2 and ccg-1 mRNA levels in the Δfrq and Δwc-1 strains (Fig. 5 B and C). These results are consistent with the osmotic sensitivity phenotypes of these strains observed on solid medium; the Δrrg-1 strain is unable to grow in conditions of osmotic stress, whereas the wild-type and clock-mutant strains display no such sensitivity (Fig. 1C and data not shown). Together, these data suggest that regulation of the OS pathway by acute osmotic shock and regulation by the circadian clock occur through different upstream pathways (Fig. 6A).
Fig. 6.
Clock control of the OS pathway may prepare cells for osmotic stress. (A) A working model of the flow of information through the OS pathway from the environment and the clock. Osmotic shock signals through the histidine kinase OS-1, and possibly other histidine kinases, to regulate the MAPK pathway and the levels of OS-2 phosphorylation and activity. Activation of OS-2 by phosphorylation results in an acute response in the downstream genes, including ccg-1, at any time of the day (red arrow). The clock signals to the same pathway at (gray arrow) or before (white arrows) RRG-1 to regulate rhythmic phosphorylation of OS-2 and rhythmic expression of at least some of the downstream genes, including ccg-1 (black arrow). (B) Model for how the clock prepares the organism for osmotic stress during the day. An osmotic shock at dawn, when ccg-1 mRNA levels are already high, would have only a small inductive effect on ccg-1 transcript levels. Alternatively, the same stress at dusk, when ccg-1 levels are low, would be expected to result in a large response and increase in ccg-1 mRNA levels. (C) Northern blots showing the levels of ccg-1 mRNA in response to the presence of 4% NaCl at DD12 or DD24 after 0 or 60 min of salt stress. The asterisks indicate treatment with distilled water instead of 4% NaCl as a control. Two autoradiograph exposures, 1 and 4 h, are shown. Ethidium-stained gels (rRNA) are shown as loading controls. This experiment was repeated two times with identical results.
Clock Control of the OS Pathway Prepares Cells for Osmotic Stress.
The finding that the clock rhythmically activates the OS pathway but is not essential for OS pathway activation by an osmotic shock suggested that the clock plays a role in preparing N. crassa cells for daily occurrences of hyperosmotic stress associated with desiccation due to sun exposure (Fig. 6 A and B). Consistent with this idea, we observed that when ccg-1 mRNA levels are already high at DD12 (subjective dawn), an osmotic shock caused only a small additional increase, whereas at DD24 (subjective dusk), an osmotic shock brought the low ccg-1 mRNA levels up to those normally seen at DD12 (Fig. 6C). These data support the hypothesis that circadian regulation of the OS pathway prepares cells for the daily hyperosmotic stress associated with desiccation by the daytime sun.
Discussion
Little is known about the output pathways from circadian oscillators in eukaryotic cells. In this study, we found that RRG-1, a component of the osmosensing signaling pathway, is also a component of an output pathway from the circadian clock that involves rhythmic activity of the p38 family MAPK OS-2. This regulation would allow the clock to prepare cells for osmotic stress that occurs at sunrise and the osmosensing system to activate the same pathway when there is unexpected osmotic stress. This mechanism may exist in all eukaryotes, because both stress-induced p38 MAPKs and circadian clocks are conserved from fungi to humans (4, 22, 23, 30–34).
The HOG pathway in S. cerevisiae controls a wide range of stress-related genes through activation of both positive and negative transcription factors by the HOG-1 MAPK (22). Many homologous genes are regulated by the OS-2 MAPK in N. crassa (33). Thus, rhythms in the activity of the OS pathway would likely influence the expression of a number of downstream genes through control of OS-2-regulated transcription factors. The delay between the induction of phospho-OS-2 (within 5 min) and the induction of ccg-1 mRNA (within 30–60 min) in response to osmotic shock (4% NaCl) is indicative of indirect regulation of ccg-1 and other genes by the OS-2 MAPK. In addition to ccg-1, the morning-specific ccg-9 and -2 genes are induced by osmotic stress and require RRG-1 for normal rhythmicity (refs. 16 and 19; SI Fig. 8). Additional loci known to be clock-regulated in N. crassa are also induced in an OS-dependent fashion in S. cerevisiae, including the genes encoding catalase-1 and alcohol dehydrogenase (13, 35). However, we believe it is unlikely that the OS pathway controls rhythmicity of all N. crassa ccgs. For example, ccg-7, encoding glyceraldehyde-3-phosphate dehydrogenase, is a morning-specific ccg that is not regulated by osmotic stress (17). Microarray experiments are currently underway to investigate which of the N. crassa ccgs are induced/repressed by osmotic shock and are rendered arrhythmic in a Δrrg-1 strain.
The Circadian Clock and Osmotic Stress Independently Regulate the OS Pathway.
The levels of total OS-2 protein are severely compromised in the Δwc-1 strain suggesting that WC-1 (perhaps through the WCC) is required for normal expression or accumulation of OS-2. However, these low levels are still sufficient for this strain to mount a response to osmotic shock; there is a robust induction of phospho-OS-2 and ccg-1 mRNA in both the Δfrq and Δwc-1 strains upon exposure to a hyperosmotic medium. These data suggest that input of information regarding the osmolarity of the environment and input of time-of-day information into this pathway occur through different upstream regulators of the OS pathway.
Compatible with FRQ's role as the negative element in the FRQ/WCC oscillator, FRQ inhibits the WC-1-dependent activation of this pathway. The levels of phosphorylated OS-2 and ccg-1 mRNA in the Δfrq strain were always higher than in wild-type strains, and the induced levels of phospo-OS-2 and ccg-1 after salt exposure were reproducibly high in the Δfrq strain as compared with wild type. The result of clock regulation of the OS pathway is a circadian rhythm in the phosphorylation or dephosphorylation, and thus activity, of the OS-2 MAPK. Consistent with posttranscriptional regulation of the activity of the OS pathway, we found in Northern blot assays that rrg-1 mRNA does not accumulate with a circadian rhythm (data not shown).
S. cerevisiae has only one sensor histidine kinase, Sln1p, and disruption of this kinase is lethal because of the constitutive activation of the HOG MAPK and buildup of glycerol in cells (36). However, the high level of active phosphorylated OS-2 observed here in Δfrq strains does not result in death on normal or hyperosmotic medium. These results accentuate several differences between the yeast HOG and N. crassa OS pathways. First, unlike yeast sln1 mutants, N. crassa os-1 histidine kinase mutants are viable under normal hypotonic conditions (24). Second, N. crassa, but not S. cerevisiae, is sensitive to phenylpyrrole fungicides that cause overactivation of the OS MAPK pathway (37, 38). Based on these data, it has been suggested that OS-1 positively regulates the OS-2 MAPK (38), whereas in S. cerevisiae, Sln1p negatively regulates the Hog1 MAPK (39). The N. crassa signaling pathways may be significantly more complex than in S. cerevisiae because of the increased number of sensor histidine kinases present in N. crassa cells (34). Moreover, activation of the OS pathway by the clock may be less than the total potential activation by osmotic shock. The high levels of ccg-1 mRNA observed in a wild-type strain at DD12, or in a Δfrq strain at all times of the day, undergo a further induction (albeit modest) during an osmotic stress (Figs. 6 and 5C, respectively). This suggests that the clock does not fully induce the OS pathway but does not rule out the possibility that there are different targets of the pathway depending on the upstream signal.
The 1-h shorter period phenotype on race tubes in Δrrg-1 strains suggests the possibility of feedback from the OS output pathway or from a downstream target(s) of this pathway to the FRQ/WCC oscillator itself. Furthermore, although normal conidial bands are not produced for the first 1–2 days in DD in the rrg-1 mutant strains, bands of aerial hyphae do occur in a rhythmic fashion. This delay in band formation is not seen when the strains are permitted to grow further down the race tube before entering DD (data not shown). Together, these observations suggest that the delay is not a circadian phenotype but is related to some other aspect of OS pathway function.
Both circadian-clock and osmotic-stress input to the OS pathway occur upstream of the MAPK cascade (involving OS-4, -5, and -2), because both forms of regulation are absent in the COP1–4 mutant and Δrrg-1 strains (Figs. 3 and 5 and data not shown; ref. 21). Precisely where the clock inputs into this pathway is not yet known. In addition to os-1, the N. crassa genome is predicted to encode 10 other hybrid histidine kinase proteins (34). It is possible that OS-1 and/or other histidine kinases could be regulated by the clock and act as circadian output kinases to transduce time-of-day information to the OS pathway.
Regulation of the OS Pathway by the Clock Prepares the Organism for Daily Hyperosmotic Conditions.
Our data suggest that regulation of the OS pathway by the clock prepares the organism for the daily hyperosmotic conditions that would be associated with desiccation by the sun in a natural environment. Up-regulation of the activity of this pathway or a subset of the components necessary for that activity may allow a preparatory response or the potential to mount a faster response at one time of day (morning) vs. another (night). Temporal regulation of the OS pathway highlights the adaptive significance of the clock, because the ability of organisms to appropriately respond to changes in osmolarity is fundamental to survival. Consistent with this idea, global gene profiling in Arabidopsis thaliana revealed that ≈68% of the ccgs overlap with genes that are differently regulated in response to osmotic and cold stresses (40).
Conserved Link Between MAPK Pathways and the Circadian Clock.
Links between Ras/ERK MAPK signaling pathways and the circadian system are known (41). For example, studies in flies and mammals have revealed effects of Ras/MAPK signaling on light-induced phase shifts of the circadian system (42–45). Additionally, the oscillator components BMAL1 (in mice) and CLK (in Drosophila) can be directly phosphorylated by MAPKs (46, 47). Although our work demonstrates circadian clock control of activity of a homolog of the p38 family of MAPKs involved in stress responses, circadian rhythms in the activity of the ERK family of MAPKs involved in growth control are also known in several systems (44, 47–55). These data suggest that control of MAPK pathways by the circadian clock may be a conserved feature of the output pathways.
Materials and Methods
Strains and Culture Conditions.
All strains used in this study are listed in SI Table 1. All strains contain the bd mutation (unless indicated otherwise), which clarifies the circadian rhythm in conidiation on growth tubes (race tubes) without otherwise affecting clock function (56). Vegetative cultures were maintained on Vogel's minimal medium (VM; 1× Vogel's salts/2% glucose) and handled according to standard procedures (57). Strains carrying the hph cassette, which confers resistance to hygromycin, were maintained on VM supplemented with 200 μg/ml hygromycin. Strains transformed with the rescue plasmid, pCJ2 (21), which contains the bar gene that confers resistance to BASTA (Bayer, Research Triangle Park, NC), were selected for on VM lacking nitrogen supplemented with 200 μg/ml BASTA/0.5% proline (as a minimal source of nitrogen).
Race tube, osmotic induction, and time-series assays were done in environmentally controlled chambers (Percival Scientific, Perry, IA). Race-tube assays were used to investigate the circadian rhythm of conidiospore development as previously described (56). Race tube media contains 1× Vogel's salts/0.1% glucose/0.5% arginine/1.5% agar. Osmotic sensitivity assays were performed on solid VM supplemented with 4% NaCl, 4% KCl, or 1 M sorbitol at 34°C in DD. Osmotic induction experiments were performed as described (18) by using liquid VM (at DD 24 unless otherwise indicated) with the following modification: 5 M NaCl was added directly to the culture medium to a final concentration of 0.7 M (4%). Time-series experiments for analysis of protein or RNA were performed as described (58) by using cultures grown in 1× Vogel's salts/2% glucose/0.5% arginine for protein extraction, and 1× Fries/0.03% glucose/0.05% arginine for RNA extraction. ccg-1 mRNA levels are much lower in the Δrrg-1 strain in the osmotic induction conditions than when they are examined in a time series, likely because of the increased glucose content of the media in the osmotic induction experiments (2% vs. 0.03%), because ccg-1 is known to be repressed in the presence of elevated glucose levels (59).
Plasmid Construction and Sequencing.
The entire rrg-1 ORF, including 271 bp 5′ and 512 bp 3′, was amplified by PCR by using genomic DNA from the wild-type or COP1–4 strain as template. The resulting 4.2-kb fragments were each cloned into the pCR-Blunt II-TOPO vector (Invitrogen, Carlsbad, CA) to produce pMV1 and pMV2, respectively. The wild-type (pMV1) and mutant rrg-1 (pMV2) locus was sequenced from these vectors. Sequencing reactions were performed by using BigDye terminator mix (Applied Biosystems, Foster City, CA) per the manufacturer's instructions and analyzed at the Gene Technology Laboratory (Institute of Developmental and Molecular Biology, Texas A&M University). Sequence analysis was performed by using Sequencher, version 4.2 (Gene Codes, Ann Arbor, MI).
Nucleic Acid Isolation, Protein Isolation, and Hybridization.
RNA isolation and Northern blot hybridization protocols have been described (60). Radioactive riboprobes were synthesized from pKL119 (ccg-1), pLMS9 (ccg-9), pLW1ΔK (ccg-2), or pMV1 (rrg-1) by using T3 or T7 polymerases, respectively, in the presence of [α-32P]UTP. Total protein isolation and Western blot hybridization were done as described in refs. 29 and 21 for detection of FRQ protein and OS-2 protein, respectively. Antibodies that recognize OS-2 total protein (anti-HOG-1, yC-20) or only phospho-OS-2 (anti-p38, Thr-180/Tyr-182) were obtained from Santa Cruz Biotechnology (Santa Cruz, CA) and Cell Signaling Technology (Danvers, MA), respectively.
Supplementary Material
Acknowledgments
We thank Drs. Richard Gomer and Teresa Lamb for comments on the manuscript and our laboratory members for general advice. This work was supported by National Institutes of Health Grant GM58529 (to D.B.-P.).
Abbreviations
- ccg
clock-controlled gene
- COP
circadian output pathway
- DD
constant darkness
- FRQ/WCC
FREQUENCY/WHITE COLLAR complex
- HOG
high-osmolarity glycerol
- OS
osmotically sensitive
- phospho-OS-2
phosphorylated OS-2 protein
- RR
response regulator.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0704900104/DC1.
References
- 1.Ouyang Y, Andersson CR, Kondo T, Golden SS, Johnson CH. Proc Natl Acad Sci USA. 1998;95:8660–8664. doi: 10.1073/pnas.95.15.8660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sharma VK. Chronobiol Int. 2003;20:901–919. doi: 10.1081/cbi-120026099. [DOI] [PubMed] [Google Scholar]
- 3.Dodd AN, Salathia N, Hall A, Kevei E, Toth R, Nagy F, Hibberd JM, Millar AJ, Webb AAR. Science. 2005;309:630–633. doi: 10.1126/science.1115581. [DOI] [PubMed] [Google Scholar]
- 4.Bell-Pedersen D, Cassone VM, Earnest DJ, Golden SS, Hardin PE, Thomas TL, Zoran MJ. Nat Rev Genet. 2005;6:544–556. doi: 10.1038/nrg1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schibler U. Prog Brain Res. 2006;153:271–282. doi: 10.1016/S0079-6123(06)53016-X. [DOI] [PubMed] [Google Scholar]
- 6.Loros JJ, Dunlap JC. Annu Rev Physiol. 2001;63:757–794. doi: 10.1146/annurev.physiol.63.1.757. [DOI] [PubMed] [Google Scholar]
- 7.Brunner M, Schafmeier T. Genes Dev. 2006;20:1061–1074. doi: 10.1101/gad.1410406. [DOI] [PubMed] [Google Scholar]
- 8.Liu Y, Bell-Pedersen D. Eukaryot Cell. 2006;5:1184–1193. doi: 10.1128/EC.00133-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dunlap JC, Loros JJ. Curr Opin Microbiol. 2006;9:579–587. doi: 10.1016/j.mib.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 10.Loros JJ, Denome SA, Dunlap JC. Science. 1989;243:385–388. doi: 10.1126/science.2563175. [DOI] [PubMed] [Google Scholar]
- 11.Bell-Pedersen D, Shinohara ML, Loros JJ, Dunlap JC. Proc Natl Acad Sci USA. 1996;93:13096–13101. doi: 10.1073/pnas.93.23.13096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu H, Nowrousian M, Kupfer D, Colot HV, Berrocal-Tito G, Lai H, Bell-Pedersen D, Roe BA, Loros JJ, Dunlap JC. Genetics. 2001;157:1057–1065. doi: 10.1093/genetics/157.3.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Correa A, Lewis ZA, Greene AV, March IJ, Gomer RH, Bell-Pedersen D. Proc Natl Acad Sci USA. 2003;100:13597–13602. doi: 10.1073/pnas.2233734100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nowrousian M, Duffield GE, Loros JJ, Dunlap JC. Genetics. 2003;164:923–933. doi: 10.1093/genetics/164.3.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pittendrigh CS, Bruce VG, Rosensweig NS, Rubin ML. Nature. 1959;184:169–170. [Google Scholar]
- 16.Shinohara ML, Loros JJ, Dunlap JC. J Biol Chem. 1998;273:446–452. doi: 10.1074/jbc.273.1.446. [DOI] [PubMed] [Google Scholar]
- 17.Bobrowicz P, Pawlak R, Correa A, Bell-Pedersen D, Ebbole DJ. Mol Microbiol. 2002;45:795–804. doi: 10.1046/j.1365-2958.2002.03052.x. [DOI] [PubMed] [Google Scholar]
- 18.Shinohara ML, Correa A, Bell-Pedersen D, Dunlap JC, Loros JJ. Eukaryot Cell. 2002;1:33–43. doi: 10.1128/EC.1.1.33-43.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vitalini MW, Morgan LW, March IJ, Bell-Pedersen D. Genetics. 2004;167:119–129. doi: 10.1534/genetics.167.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindgren KM. Hanover, NH: Dartmouth College; 1994. PhD thesis. [Google Scholar]
- 21.Jones CA, Greer-Phillips SA, Borkovich KA. Mol Biol Cell. 2007;18:2123–2136. doi: 10.1091/mbc.E06-03-0226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hohmann S. Microbiol Mol Biol Rev. 2002;66:300–372. doi: 10.1128/MMBR.66.2.300-372.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ikner A, Shiozaki K. Mutat Res. 2005;569:13–27. doi: 10.1016/j.mrfmmm.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 24.Schumacher MM, Enderlin CS, Selitrennikoff CP. Curr Microbiol. 1997;34:340–347. doi: 10.1007/s002849900193. [DOI] [PubMed] [Google Scholar]
- 25.Fujimura M, Ochiai N, Oshima M, Motoyama T, Ichiishi A, Usami R, Horikoshi K, Yamaguchi I. Biosci Biotechnol Biochem. 2003;67:186–191. doi: 10.1271/bbb.67.186. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y, Lamm R, Pillonel C, Lam S, Xu JR. Appl Environ Microbiol. 2002;68:532–538. doi: 10.1128/AEM.68.2.532-538.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee K, Dunlap JC, Loros JJ. Genetics. 2003;163:103–114. doi: 10.1093/genetics/163.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vitalini MW, de Paula RM, Park WD, Bell-Pedersen D. J Biol Rhythms. 2006;21:432–444. doi: 10.1177/0748730406294396. [DOI] [PubMed] [Google Scholar]
- 29.Garceau NY, Liu Y, Loros JJ, Dunlap JC. Cell. 1997;89:469–476. doi: 10.1016/s0092-8674(00)80227-5. [DOI] [PubMed] [Google Scholar]
- 30.Hardin PE. Curr Opin Neurobiol. 2006;16:686–692. doi: 10.1016/j.conb.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 31.Zarubin T, Han J. Cell Res. 2005;15:11–18. doi: 10.1038/sj.cr.7290257. [DOI] [PubMed] [Google Scholar]
- 32.Qi M, Elion EA. J Cell Sci. 2005;118:3569–3572. doi: 10.1242/jcs.02470. [DOI] [PubMed] [Google Scholar]
- 33.Noguchi R, Banno S, Ichikawa R, Fukumori F, Ichiishi A, Kimura M, Yamaguchi I, Fujimura M. Fungal Genet Biol. 2006;44:208–218. doi: 10.1016/j.fgb.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 34.Borkovich KA, Alex LA, Yarden O, Freitag M, Turner GE, Read ND, Seiler S, Bell-Pedersen D, Paietta J, Plesofsky N, et al. Microbiol Mol Biol Rev. 2004;68:1–108. doi: 10.1128/MMBR.68.1.1-108.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rep M, Krantz M, Thevelein JM, Hohmann S. J Biol Chem. 2000;275:8290–8300. doi: 10.1074/jbc.275.12.8290. [DOI] [PubMed] [Google Scholar]
- 36.Maeda T, Wurgler-Murphy SM, Saito H. Nature. 1994;369:242–245. doi: 10.1038/369242a0. [DOI] [PubMed] [Google Scholar]
- 37.Kojima K, Takano Y, Yoshimi A, Tanaka C, Kikuchi T, Okuno T. Mol Microbiol. 2004;53:1785–1796. doi: 10.1111/j.1365-2958.2004.04244.x. [DOI] [PubMed] [Google Scholar]
- 38.Yoshimi A, Kojima K, Takano Y, Tanaka C. Eukaryot Cell. 2005;4:1820–1828. doi: 10.1128/EC.4.11.1820-1828.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Posas F, Wurgler-Murphy SM, Maeda T, Witten EA, Thai TC, Saito H. Cell. 1996;86:865–875. doi: 10.1016/s0092-8674(00)80162-2. [DOI] [PubMed] [Google Scholar]
- 40.Kreps JA, Wu Y, Chang HS, Zhu T, Wang X, Harper JF. Plant Physiol. 2002;130:2129–2141. doi: 10.1104/pp.008532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coogan AN, Piggins HD. J Neurochem. 2004;90:769–775. doi: 10.1111/j.1471-4159.2004.02554.x. [DOI] [PubMed] [Google Scholar]
- 42.Akashi M, Nishida E. Genes Dev. 2000;14:645–649. [PMC free article] [PubMed] [Google Scholar]
- 43.Butcher GQ, Dziema H, Collamore M, Burgoon PW, Obrietan K. J Biol Chem. 2002;277:29519–29525. doi: 10.1074/jbc.M203301200. [DOI] [PubMed] [Google Scholar]
- 44.Hayashi Y, Sanada K, Fukada Y. FEBS Lett. 2001;491:71–75. doi: 10.1016/s0014-5793(01)02153-6. [DOI] [PubMed] [Google Scholar]
- 45.Hayashi Y, Sanada K, Hirota T, Shimizu F, Fukada Y. J Biol Chem. 2003;278:25166–25171. doi: 10.1074/jbc.M212726200. [DOI] [PubMed] [Google Scholar]
- 46.Sanada K, Okano T, Fukada Y. J Biol Chem. 2002;277:267–271. doi: 10.1074/jbc.M107850200. [DOI] [PubMed] [Google Scholar]
- 47.Weber F, Hung HC, Maurer C, Kay SA. J Neurochem. 2006;98:248–257. doi: 10.1111/j.1471-4159.2006.03865.x. [DOI] [PubMed] [Google Scholar]
- 48.Williams JA, Su HS, Bernards A, Field J, Sehgal A. Science. 2001;293:2251–2256. doi: 10.1126/science.1063097. [DOI] [PubMed] [Google Scholar]
- 49.Shim HS, Kim H, Lee J, Son GH, Cho S, Oh TH, Kang SH, Seen DS, Lee KH, Kim K. EMBO Rep. 2007;8:366–371. doi: 10.1038/sj.embor.7400920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sanada K, Hayashi Y, Harada Y, Okano T, Fukada Y. J Neurosci. 2000;20:986–991. doi: 10.1523/JNEUROSCI.20-03-00986.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Harada Y, Sanada K, Fukada Y. J Biol Chem. 2000;275:37078–37085. doi: 10.1074/jbc.M004706200. [DOI] [PubMed] [Google Scholar]
- 52.Ko GY, Ko ML, Dryer SE. Neuron. 2001;29:255–266. doi: 10.1016/s0896-6273(01)00195-7. [DOI] [PubMed] [Google Scholar]
- 53.Nakaya M, Sanada K, Fukada Y. Biochem Biophys Res Commun. 2003;305:494–501. doi: 10.1016/s0006-291x(03)00791-5. [DOI] [PubMed] [Google Scholar]
- 54.Obrietan K, Impey S, Storm DR. Nat Neurosci. 1998;1:693–700. doi: 10.1038/3695. [DOI] [PubMed] [Google Scholar]
- 55.Pizzio GA, Hainich EC, Ferreyra GA, Coso OA, Golombek DA. NeuroReport. 2003;14:1417–1419. doi: 10.1097/00001756-200308060-00002. [DOI] [PubMed] [Google Scholar]
- 56.Sargent ML, Kaltenborn SH. Plant Physiol. 1972;50:171–175. doi: 10.1104/pp.50.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Davis RL, DeSerres D. Methods Enzymol. 1970;27A:79–143. [Google Scholar]
- 58.Correa A, Bell-Pedersen D. Eukaryot Cell. 2002;1:273–280. doi: 10.1128/EC.1.2.273-280.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McNally MT, Free SJ. Curr Genet. 1988;14:545–551. doi: 10.1007/BF00434079. [DOI] [PubMed] [Google Scholar]
- 60.Bell-Pedersen D, Dunlap JC, Loros JJ. Mol Cell Biol. 1996;16:513–521. doi: 10.1128/mcb.16.2.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aronson BD, Johnson KA, Dunlap JC. Proc Natl Acad Sci USA. 1994;91:7683–7687. doi: 10.1073/pnas.91.16.7683. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.