Abstract
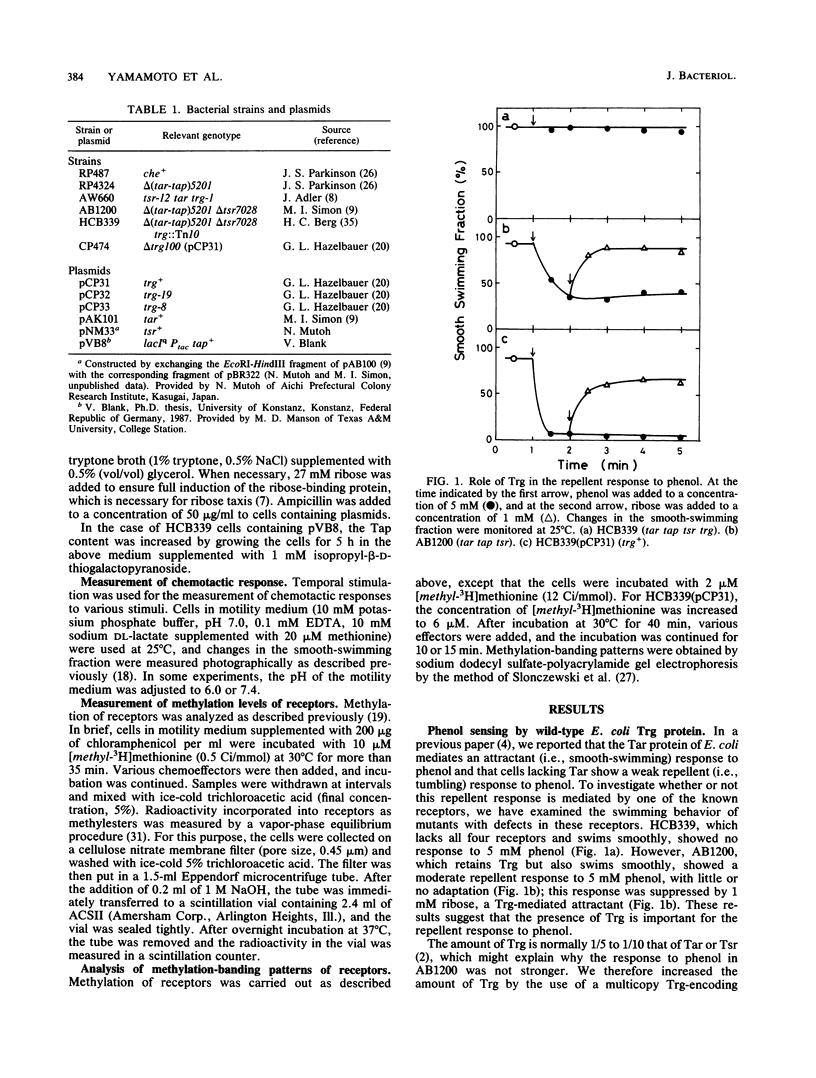
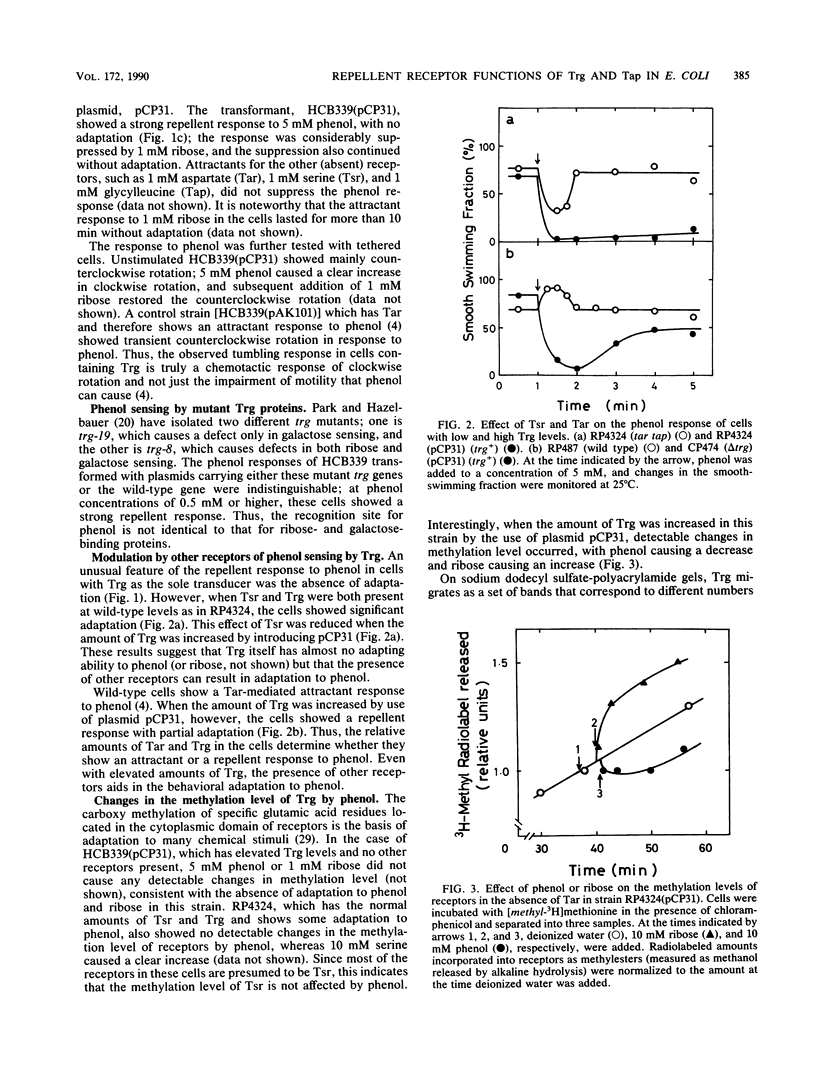
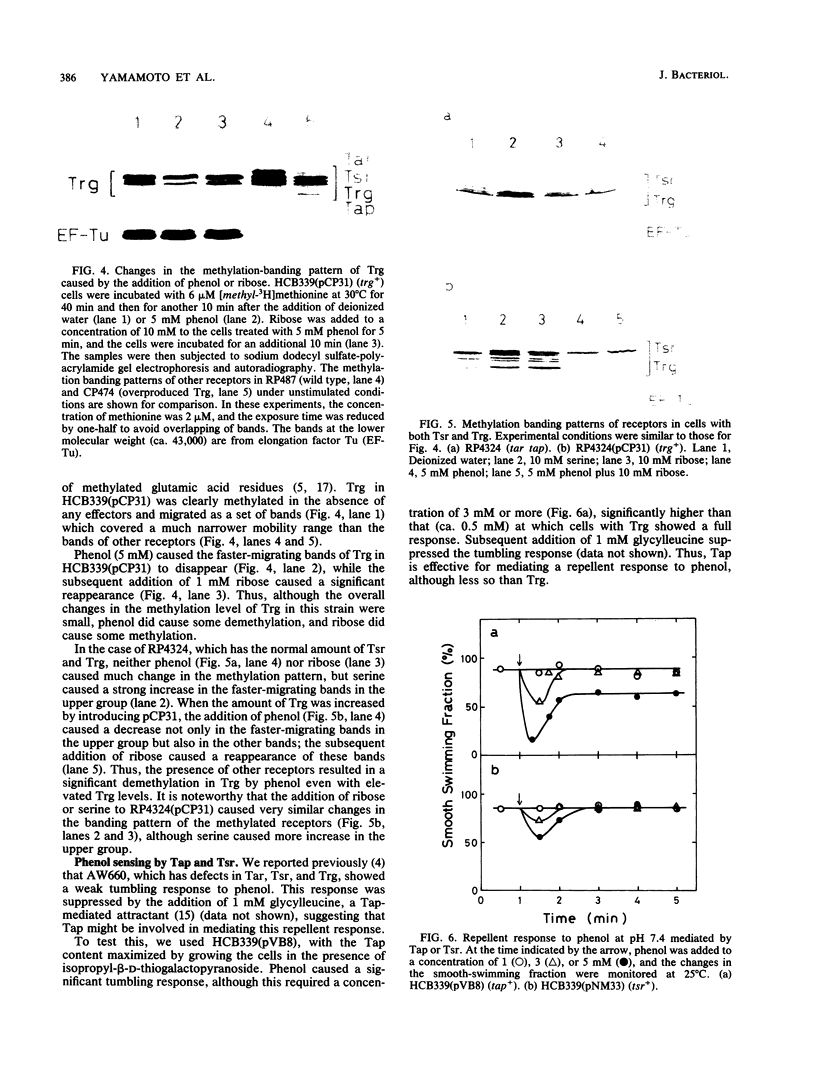
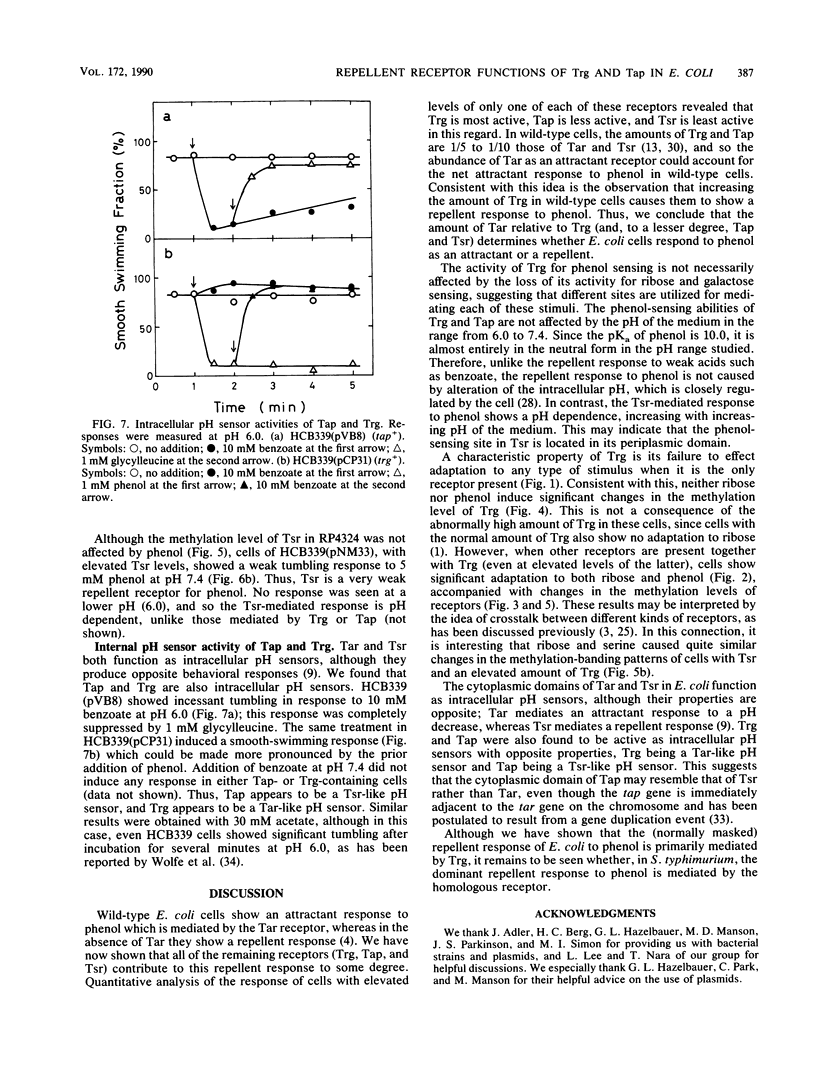
The chemoreceptors responsible for the repellent response of Escherichia coli to phenol were investigated. In the absence of all four known methyl-accepting chemoreceptors (Tar, Tsr, Trg, and Tap), cells showed no response to phenol. However, when Trg, which mediates the attractant response to ribose and galactose, was introduced via a plasmid, the cells acquired a repellent response to phenol. About 1 mM phenol induced a clear repellent response; this response was suppressed by 1 mM ribose. Thus, Trg mediates the repellent response to phenol. Mutant Trg proteins with altered sensing for ribose and galactose showed a normal response to phenol, indicating that the interaction site for phenol differs from that for the ribose- and galactose-binding proteins. Tap, which mediates the attractant response to dipeptides, mediated a weaker repellent response to phenol. Tsr, which mediates the attractant response to serine, mediated an even weaker response to phenol. Trg and Tap were also found to function as intracellular pH sensors. Upon a pH decrease, Trg mediated an attractant response, whereas Tap mediated a repellent response. These results indicate that all the receptors in E. coli have dual functions, mediating both attractant and repellent responses.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Hazelbauer G. L., Engström P. Multiple forms of methyl-accepting chemotaxis proteins distinguished by a factor in addition to multiple methylation. J Bacteriol. 1981 Jan;145(1):35–42. doi: 10.1128/jb.145.1.35-42.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazelbauer G. L., Engström P. Parallel pathways for transduction of chemotactic signals in Escherichia coli. Nature. 1980 Jan 3;283(5742):98–100. doi: 10.1038/283098a0. [DOI] [PubMed] [Google Scholar]
- Hazelbauer G. L., Park C., Nowlin D. M. Adaptational "crosstalk" and the crucial role of methylation in chemotactic migration by Escherichia coli. Proc Natl Acad Sci U S A. 1989 Mar;86(5):1448–1452. doi: 10.1073/pnas.86.5.1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imae Y., Oosawa K., Mizuno T., Kihara M., Macnab R. M. Phenol: a complex chemoeffector in bacterial chemotaxis. J Bacteriol. 1987 Jan;169(1):371–379. doi: 10.1128/jb.169.1.371-379.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehry M. R., Engström P., Dahlquist F. W., Hazelbauer G. L. Multiple covalent modifications of Trg, a sensory transducer of Escherichia coli. J Biol Chem. 1983 Apr 25;258(8):5050–5055. [PubMed] [Google Scholar]
- Kihara M., Macnab R. M. Cytoplasmic pH mediates pH taxis and weak-acid repellent taxis of bacteria. J Bacteriol. 1981 Mar;145(3):1209–1221. doi: 10.1128/jb.145.3.1209-1221.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koman A., Harayama S., Hazelbauer G. L. Relation of chemotactic response to the amount of receptor: evidence for different efficiencies of signal transduction. J Bacteriol. 1979 Jun;138(3):739–747. doi: 10.1128/jb.138.3.739-747.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondoh H., Ball C. B., Adler J. Identification of a methyl-accepting chemotaxis protein for the ribose and galactose chemoreceptors of Escherichia coli. Proc Natl Acad Sci U S A. 1979 Jan;76(1):260–264. doi: 10.1073/pnas.76.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krikos A., Conley M. P., Boyd A., Berg H. C., Simon M. I. Chimeric chemosensory transducers of Escherichia coli. Proc Natl Acad Sci U S A. 1985 Mar;82(5):1326–1330. doi: 10.1073/pnas.82.5.1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krikos A., Mutoh N., Boyd A., Simon M. I. Sensory transducers of E. coli are composed of discrete structural and functional domains. Cell. 1983 Jun;33(2):615–622. doi: 10.1016/0092-8674(83)90442-7. [DOI] [PubMed] [Google Scholar]
- Lederberg J. Linear Inheritance in Transductional Clones. Genetics. 1956 Nov;41(6):845–871. doi: 10.1093/genetics/41.6.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee L., Mizuno T., Imae Y. Thermosensing properties of Escherichia coli tsr mutants defective in serine chemoreception. J Bacteriol. 1988 Oct;170(10):4769–4774. doi: 10.1128/jb.170.10.4769-4774.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoil C., Beckwith J. A genetic approach to analyzing membrane protein topology. Science. 1986 Sep 26;233(4771):1403–1408. doi: 10.1126/science.3529391. [DOI] [PubMed] [Google Scholar]
- Manson M. D., Blank V., Brade G., Higgins C. F. Peptide chemotaxis in E. coli involves the Tap signal transducer and the dipeptide permease. Nature. 1986 May 15;321(6067):253–256. doi: 10.1038/321253a0. [DOI] [PubMed] [Google Scholar]
- Mizuno T., Mutoh N., Panasenko S. M., Imae Y. Acquisition of maltose chemotaxis in Salmonella typhimurium by the introduction of the Escherichia coli chemosensory transducer gene. J Bacteriol. 1986 Mar;165(3):890–895. doi: 10.1128/jb.165.3.890-895.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowlin D. M., Bollinger J., Hazelbauer G. L. Site-directed mutations altering methyl-accepting residues of a sensory transducer protein. Proteins. 1988;3(2):102–112. doi: 10.1002/prot.340030205. [DOI] [PubMed] [Google Scholar]
- Oosawa K., Imae Y. Demethylation of methyl-accepting chemotaxis proteins in Escherichia coli induced by the repellents glycerol and ethylene glycol. J Bacteriol. 1984 Feb;157(2):576–581. doi: 10.1128/jb.157.2.576-581.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oosawa K., Imae Y. Glycerol and ethylene glycol: members of a new class of repellents of Escherichia coli chemotaxis. J Bacteriol. 1983 Apr;154(1):104–112. doi: 10.1128/jb.154.1.104-112.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C., Hazelbauer G. L. Mutations specifically affecting ligand interaction of the Trg chemosensory transducer. J Bacteriol. 1986 Jul;167(1):101–109. doi: 10.1128/jb.167.1.101-109.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson J. S. Protein phosphorylation in bacterial chemotaxis. Cell. 1988 Apr 8;53(1):1–2. doi: 10.1016/0092-8674(88)90478-3. [DOI] [PubMed] [Google Scholar]
- Repaske D. R., Adler J. Change in intracellular pH of Escherichia coli mediates the chemotactic response to certain attractants and repellents. J Bacteriol. 1981 Mar;145(3):1196–1208. doi: 10.1128/jb.145.3.1196-1208.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo A. F., Koshland D. E., Jr Identification of the tip-encoded receptor in bacterial sensing. J Bacteriol. 1986 Jan;165(1):276–282. doi: 10.1128/jb.165.1.276-282.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo A. F., Koshland D. E., Jr Separation of signal transduction and adaptation functions of the aspartate receptor in bacterial sensing. Science. 1983 Jun 3;220(4601):1016–1020. doi: 10.1126/science.6302843. [DOI] [PubMed] [Google Scholar]
- Sanders D. A., Koshland D. E., Jr Receptor interactions through phosphorylation and methylation pathways in bacterial chemotaxis. Proc Natl Acad Sci U S A. 1988 Nov;85(22):8425–8429. doi: 10.1073/pnas.85.22.8425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slocum M. K., Parkinson J. S. Genetics of methyl-accepting chemotaxis proteins in Escherichia coli: organization of the tar region. J Bacteriol. 1983 Aug;155(2):565–577. doi: 10.1128/jb.155.2.565-577.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slonczewski J. L., Macnab R. M., Alger J. R., Castle A. M. Effects of pH and repellent tactic stimuli on protein methylation levels in Escherichia coli. J Bacteriol. 1982 Oct;152(1):384–399. doi: 10.1128/jb.152.1.384-399.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slonczewski J. L., Rosen B. P., Alger J. R., Macnab R. M. pH homeostasis in Escherichia coli: measurement by 31P nuclear magnetic resonance of methylphosphonate and phosphate. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6271–6275. doi: 10.1073/pnas.78.10.6271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer M. S., Goy M. F., Adler J. Protein methylation in behavioural control mechanisms and in signal transduction. Nature. 1979 Jul 26;280(5720):279–284. doi: 10.1038/280279a0. [DOI] [PubMed] [Google Scholar]
- Stock J. B., Clarke S., Koshland D. E., Jr The protein carboxylmethyltransferase involved in Escherichia coli and Salmonella typhimurium chemotaxis. Methods Enzymol. 1984;106:310–321. doi: 10.1016/0076-6879(84)06031-6. [DOI] [PubMed] [Google Scholar]
- Tso W. W., Adler J. Negative chemotaxis in Escherichia coli. J Bacteriol. 1974 May;118(2):560–576. doi: 10.1128/jb.118.2.560-576.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E. A., Mowry K. L., Clegg D. O., Koshland D. E., Jr Tandem duplication and multiple functions of a receptor gene in bacterial chemotaxis. J Biol Chem. 1982 May 10;257(9):4673–4676. [PubMed] [Google Scholar]
- Wolfe A. J., Conley M. P., Berg H. C. Acetyladenylate plays a role in controlling the direction of flagellar rotation. Proc Natl Acad Sci U S A. 1988 Sep;85(18):6711–6715. doi: 10.1073/pnas.85.18.6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe A. J., Conley M. P., Kramer T. J., Berg H. C. Reconstitution of signaling in bacterial chemotaxis. J Bacteriol. 1987 May;169(5):1878–1885. doi: 10.1128/jb.169.5.1878-1885.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]