Abstract
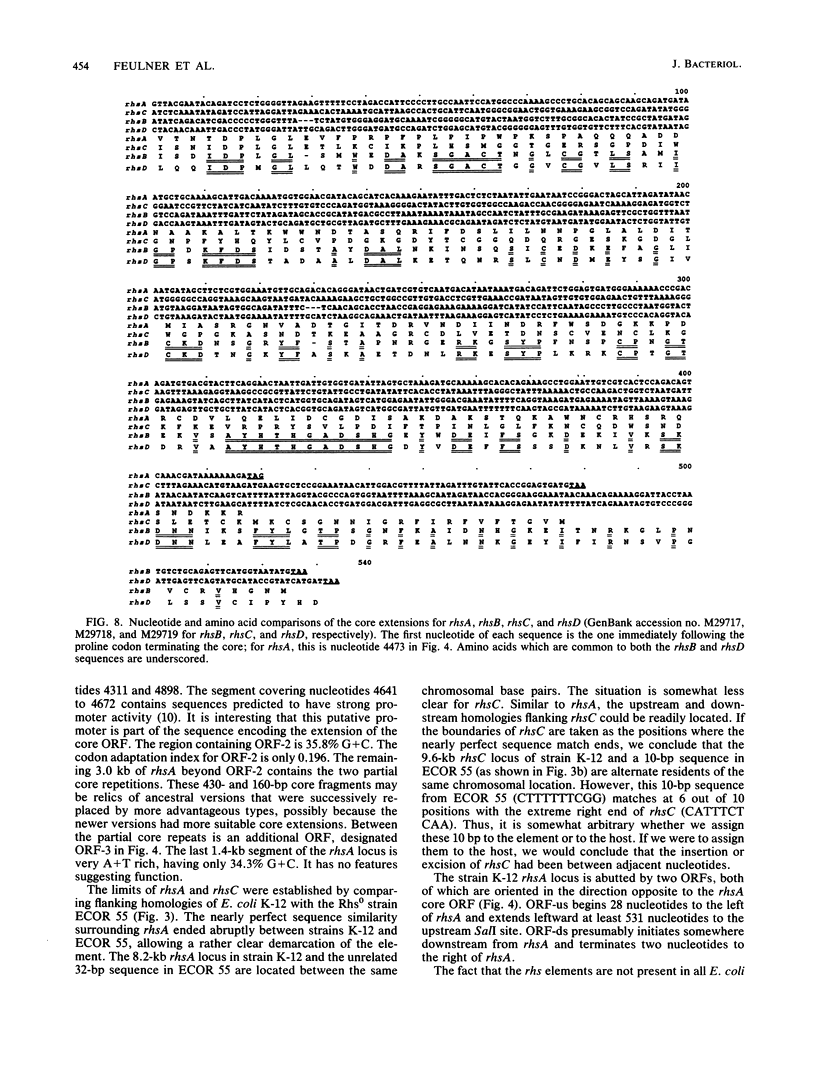
The complete nucleotide sequence of the rhsA locus and selected portions of other members of the rhs multigene family of Escherichia coli K-12 have been determined. A definition of the limits of the rhsA and rhsC loci was established by comparing sequences from E. coli K-12 with sequences from an independent E. coli isolate whose DNA contains no homology to the rhs core. This comparison showed that rhsA comprises 8,249 base pairs (bp) in strain K-12 and that the Rhs0 strain, instead, contains an unrelated 32-bp sequence. Similarly, the K-12 rhsC locus is 9.6 kilobases in length and a 10-bp sequence resides at its location in the Rhs0 strain. The rhsA core, the highly conserved portion shared by all rhs loci, comprises a single open reading frame (ORF) 3,714 bp in length. The nucleotide sequence of the core ORF predicts an extremely hydrophilic 141-kilodalton peptide containing 28 repeats of a motif whose consensus is GxxxRYxYDxxGRL(I or T). One of the most novel aspects of the rhs family is the extension of the core ORF into the divergent adjacent region. Core extensions of rhsA, rhsB, rhsC, and rhsD add 139, 173, 159, and 177 codons to the carboxy termini of the respective core ORFs. For rhsA, the extended core protein would have a molecular mass of 156 kilodaltons. Core extensions of rhsB and rhsD are related, exhibiting 50.3% conservation of the predicted amino acid sequence. However, comparison of the core extensions of rhsA and rhsC at both the nucleotide and the predicted amino acid level reveals that each is highly divergent from the other three rhs loci. The highly divergent portion of the core extension is joined to the highly conserved core by a nine-codon segment of intermediate conservation. The rhsA and rhsC loci both contain partial repetitions of the core downstream from their primary cores. The question of whether the rhs loci should be considered accessory genetic elements is discussed but not resolved.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bolivar F. Construction and characterization of new cloning vehicles. III. Derivatives of plasmid pBR322 carrying unique Eco RI sites for selection of Eco RI generated recombinant DNA molecules. Gene. 1978 Oct;4(2):121–136. doi: 10.1016/0378-1119(78)90025-2. [DOI] [PubMed] [Google Scholar]
- Brody H., Greener A., Hill C. W. Excision and reintegration of the Escherichia coli K-12 chromosomal element e14. J Bacteriol. 1985 Mar;161(3):1112–1117. doi: 10.1128/jb.161.3.1112-1117.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell A. Evolutionary significance of accessory DNA elements in bacteria. Annu Rev Microbiol. 1981;35:55–83. doi: 10.1146/annurev.mi.35.100181.000415. [DOI] [PubMed] [Google Scholar]
- Capage M., Hill C. W. Preferential unequal recombination in the glyS region of the Escherichia coli chromosome. J Mol Biol. 1979 Jan 5;127(1):73–87. doi: 10.1016/0022-2836(79)90460-1. [DOI] [PubMed] [Google Scholar]
- Casadaban M. J., Chou J., Cohen S. N. In vitro gene fusions that join an enzymatically active beta-galactosidase segment to amino-terminal fragments of exogenous proteins: Escherichia coli plasmid vectors for the detection and cloning of translational initiation signals. J Bacteriol. 1980 Aug;143(2):971–980. doi: 10.1128/jb.143.2.971-980.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban M. J., Martinez-Arias A., Shapira S. K., Chou J. Beta-galactosidase gene fusions for analyzing gene expression in escherichia coli and yeast. Methods Enzymol. 1983;100:293–308. doi: 10.1016/0076-6879(83)00063-4. [DOI] [PubMed] [Google Scholar]
- Felmlee T., Pellett S., Welch R. A. Nucleotide sequence of an Escherichia coli chromosomal hemolysin. J Bacteriol. 1985 Jul;163(1):94–105. doi: 10.1128/jb.163.1.94-105.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grindley N. D., Joyce C. M. Genetic and DNA sequence analysis of the kanamycin resistance transposon Tn903. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7176–7180. doi: 10.1073/pnas.77.12.7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley D. K., McClure W. R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983 Apr 25;11(8):2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R. J., Capage M., Hill C. W. A repetitive DNA sequence, rhs, responsible for duplications within the Escherichia coli K-12 chromosome. J Mol Biol. 1984 Jul 25;177(1):1–18. doi: 10.1016/0022-2836(84)90054-8. [DOI] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Ochman H., Selander R. K. Standard reference strains of Escherichia coli from natural populations. J Bacteriol. 1984 Feb;157(2):690–693. doi: 10.1128/jb.157.2.690-693.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen G. B., Stockwell P. A., Hill D. F. Messenger RNA recognition in Escherichia coli: a possible second site of interaction with 16S ribosomal RNA. EMBO J. 1988 Dec 1;7(12):3957–3962. doi: 10.1002/j.1460-2075.1988.tb03282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petes T. D., Hill C. W. Recombination between repeated genes in microorganisms. Annu Rev Genet. 1988;22:147–168. doi: 10.1146/annurev.ge.22.120188.001051. [DOI] [PubMed] [Google Scholar]
- Prentki P., Karch F., Iida S., Meyer J. The plasmid cloning vector pBR325 contains a 482 base-pair-long inverted duplication. Gene. 1981 Sep;14(4):289–299. doi: 10.1016/0378-1119(81)90161-x. [DOI] [PubMed] [Google Scholar]
- Sadosky A. B., Davidson A., Lin R. J., Hill C. W. rhs gene family of Escherichia coli K-12. J Bacteriol. 1989 Feb;171(2):636–642. doi: 10.1128/jb.171.2.636-642.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A., Hack A. M., Rupp W. D. Simple method for identification of plasmid-coded proteins. J Bacteriol. 1979 Jan;137(1):692–693. doi: 10.1128/jb.137.1.692-693.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. M., Li W. H. The codon Adaptation Index--a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987 Feb 11;15(3):1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signäs C., Raucci G., Jönsson K., Lindgren P. E., Anantharamaiah G. M., Hök M., Lindberg M. Nucleotide sequence of the gene for a fibronectin-binding protein from Staphylococcus aureus: use of this peptide sequence in the synthesis of biologically active peptides. Proc Natl Acad Sci U S A. 1989 Jan;86(2):699–703. doi: 10.1073/pnas.86.2.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. Membrane proteins: the amino acid composition of membrane-penetrating segments. Eur J Biochem. 1981 Nov;120(2):275–278. doi: 10.1111/j.1432-1033.1981.tb05700.x. [DOI] [PubMed] [Google Scholar]
- von Heijne G. Signal sequences. The limits of variation. J Mol Biol. 1985 Jul 5;184(1):99–105. doi: 10.1016/0022-2836(85)90046-4. [DOI] [PubMed] [Google Scholar]



