Abstract
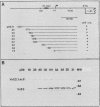
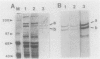
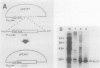
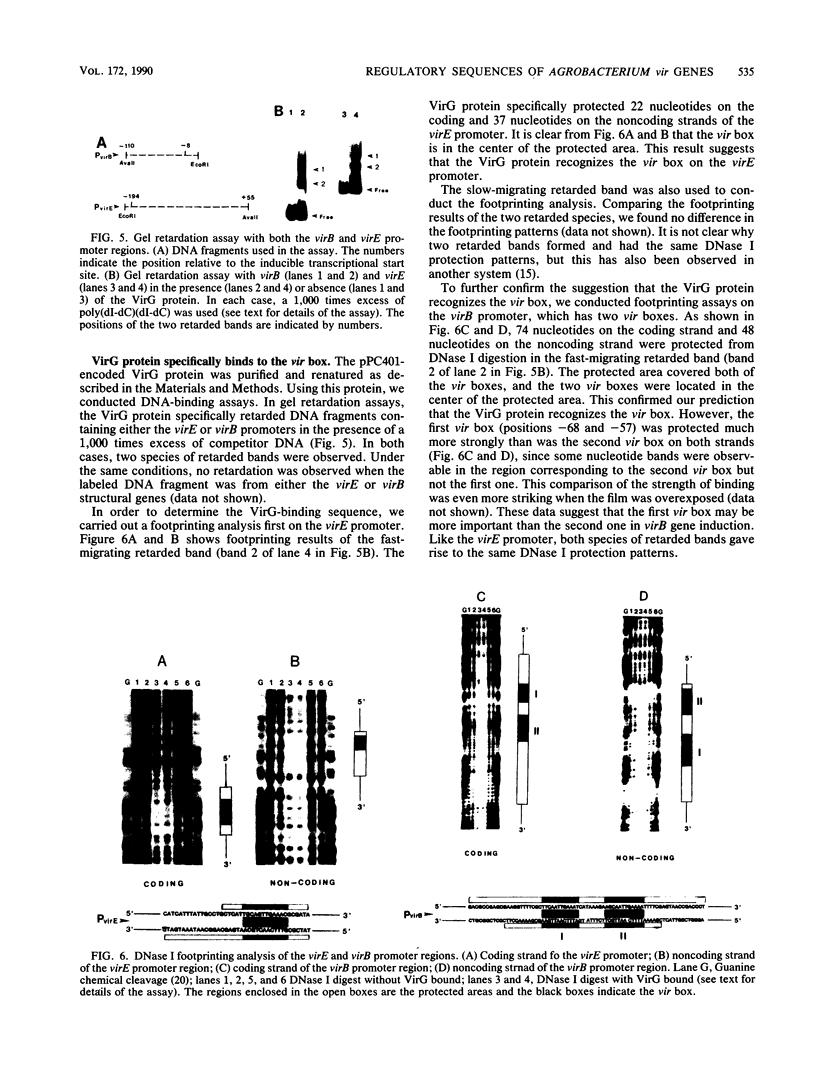
Virulence genes of Agrobacterium tumefaciens are induced in parallel in the presence of plant phenolic compounds such as acetosyringone and the two regulatory vir genes virA and virG. In this study we identified a cis-acting regulatory sequence in the 5'-noncoding region of the virE operon that is essential for this activation. To do this, we constructed a series of deletion mutants by using exonuclease Bal 31. Western blot (immunoblot) analysis showed that the 70 base pairs upstream of the transcriptional start site were sufficient for full virE gene induction. A conserved dodecadeoxynucleotide sequence (vir box), which was previously identified in the nontranscribed sequences of all vir genes, was located at 5' end of the minimum required promoter sequence. Deletion of this vir box only completely abolished induction of the virE gene. This demonstrates that the vir box functions as an upstream regulatory sequence. To study the role of the VirG protein in the activation process, we overproduced the native-sized VirG protein in Escherichia coli by fusing the lacZ' start codon ATG with the second virG codon AAA using site-directed mutagenesis. The VirG protein was purified and renatured from E. coli and was shown to bind to a specific sequence in two vir gene promoters. Footprinting analysis of the virE and virB promoters identified the 12-base-pair vir box as the VirG-binding core sequence.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Banerji J., Rusconi S., Schaffner W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981 Dec;27(2 Pt 1):299–308. doi: 10.1016/0092-8674(81)90413-x. [DOI] [PubMed] [Google Scholar]
- Birkmann A., Böck A. Characterization of a cis regulatory DNA element necessary for formate induction of the formate dehydrogenase gene (fdhF) of Escherichia coli. Mol Microbiol. 1989 Feb;3(2):187–195. doi: 10.1111/j.1365-2958.1989.tb01807.x. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Christie P. J., Ward J. E., Winans S. C., Nester E. W. The Agrobacterium tumefaciens virE2 gene product is a single-stranded-DNA-binding protein that associates with T-DNA. J Bacteriol. 1988 Jun;170(6):2659–2667. doi: 10.1128/jb.170.6.2659-2667.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citovsky V., DE Vos G., Zambryski P. Single-Stranded DNA Binding Protein Encoded by the virE Locus of Agrobacterium tumefaciens. Science. 1988 Apr 22;240(4851):501–504. doi: 10.1126/science.240.4851.501. [DOI] [PubMed] [Google Scholar]
- Close T. J., Zaitlin D., Kado C. I. Design and development of amplifiable broad-host-range cloning vectors: analysis of the vir region of Agrobacterium tumefaciens plasmid pTiC58. Plasmid. 1984 Sep;12(2):111–118. doi: 10.1016/0147-619x(84)90057-x. [DOI] [PubMed] [Google Scholar]
- Das A., Pazour G. J. Delineation of the regulatory region sequences of Agrobacterium tumefaciens virB operon. Nucleic Acids Res. 1989 Jun 26;17(12):4541–4550. doi: 10.1093/nar/17.12.4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A., Stachel S., Ebert P., Allenza P., Montoya A., Nester E. Promoters of Agrobacterium tumefaciens Ti-plasmid virulence genes. Nucleic Acids Res. 1986 Feb 11;14(3):1355–1364. doi: 10.1093/nar/14.3.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditta G., Stanfield S., Corbin D., Helinski D. R. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferro-Luzzi Ames G., Nikaido K. Nitrogen regulation in Salmonella typhimurium. Identification of an ntrC protein-binding site and definition of a consensus binding sequence. EMBO J. 1985 Feb;4(2):539–547. doi: 10.1002/j.1460-2075.1985.tb03662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hager D. A., Burgess R. R. Elution of proteins from sodium dodecyl sulfate-polyacrylamide gels, removal of sodium dodecyl sulfate, and renaturation of enzymatic activity: results with sigma subunit of Escherichia coli RNA polymerase, wheat germ DNA topoisomerase, and other enzymes. Anal Biochem. 1980 Nov 15;109(1):76–86. doi: 10.1016/0003-2697(80)90013-5. [DOI] [PubMed] [Google Scholar]
- Jin S., Roitsch T., Ankenbauer R. G., Gordon M. P., Nester E. W. The VirA protein of Agrobacterium tumefaciens is autophosphorylated and is essential for vir gene regulation. J Bacteriol. 1990 Feb;172(2):525–530. doi: 10.1128/jb.172.2.525-530.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanemoto R. H., Powell A. T., Akiyoshi D. E., Regier D. A., Kerstetter R. A., Nester E. W., Hawes M. C., Gordon M. P. Nucleotide sequence and analysis of the plant-inducible locus pinF from Agrobacterium tumefaciens. J Bacteriol. 1989 May;171(5):2506–2512. doi: 10.1128/jb.171.5.2506-2512.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Landolfi N. F., Capra J. D., Tucker P. W. Protein-nucleotide contacts in the immunoglobulin heavy-chain promoter region. Proc Natl Acad Sci U S A. 1987 Jun;84(11):3851–3855. doi: 10.1073/pnas.84.11.3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroux B., Yanofsky M. F., Winans S. C., Ward J. E., Ziegler S. F., Nester E. W. Characterization of the virA locus of Agrobacterium tumefaciens: a transcriptional regulator and host range determinant. EMBO J. 1987 Apr;6(4):849–856. doi: 10.1002/j.1460-2075.1987.tb04830.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda S., Ozawa Y., Mizuno T., Mizushima S. Stereospecific positioning of the cis-acting sequence with respect to the canonical promoter is required for activation of the ompC gene by a positive regulator, OmpR, in Escherichia coli. J Mol Biol. 1988 Aug 5;202(3):433–441. doi: 10.1016/0022-2836(88)90276-8. [DOI] [PubMed] [Google Scholar]
- Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem. 1987 Jul 25;262(21):10035–10038. [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Mitchell P. J., Tjian R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science. 1989 Jul 28;245(4916):371–378. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- Mizuno T., Kato M., Jo Y. L., Mizushima S. Interaction of OmpR, a positive regulator, with the osmoregulated ompC and ompF genes of Escherichia coli. Studies with wild-type and mutant OmpR proteins. J Biol Chem. 1988 Jan 15;263(2):1008–1012. [PubMed] [Google Scholar]
- Murray R. G. A structured life. Annu Rev Microbiol. 1988;42:1–34. doi: 10.1146/annurev.mi.42.100188.000245. [DOI] [PubMed] [Google Scholar]
- Ninfa A. J., Magasanik B. Covalent modification of the glnG product, NRI, by the glnL product, NRII, regulates the transcription of the glnALG operon in Escherichia coli. Proc Natl Acad Sci U S A. 1986 Aug;83(16):5909–5913. doi: 10.1073/pnas.83.16.5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popham D. L., Szeto D., Keener J., Kustu S. Function of a bacterial activator protein that binds to transcriptional enhancers. Science. 1989 Feb 3;243(4891):629–635. doi: 10.1126/science.2563595. [DOI] [PubMed] [Google Scholar]
- Powell B. S., Rogowsky P. M., Kado C. I. virG of Agrobacterium tumefaciens plasmid pTiC58 encodes a DNA-binding protein. Mol Microbiol. 1989 Mar;3(3):411–419. doi: 10.1111/j.1365-2958.1989.tb00186.x. [DOI] [PubMed] [Google Scholar]
- Powell G. K., Hommes N. G., Kuo J., Castle L. A., Morris R. O. Inducible expression of cytokinin biosynthesis in Agrobacterium tumefaciens by plant phenolics. Mol Plant Microbe Interact. 1988 Jul-Aug;1(6):235–242. doi: 10.1094/mpmi-1-235. [DOI] [PubMed] [Google Scholar]
- Ronson C. W., Nixon B. T., Ausubel F. M. Conserved domains in bacterial regulatory proteins that respond to environmental stimuli. Cell. 1987 Jun 5;49(5):579–581. doi: 10.1016/0092-8674(87)90530-7. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachel S. E., Nester E. W. The genetic and transcriptional organization of the vir region of the A6 Ti plasmid of Agrobacterium tumefaciens. EMBO J. 1986 Jul;5(7):1445–1454. doi: 10.1002/j.1460-2075.1986.tb04381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachel S. E., Zambryski P. C. virA and virG control the plant-induced activation of the T-DNA transfer process of A. tumefaciens. Cell. 1986 Aug 1;46(3):325–333. doi: 10.1016/0092-8674(86)90653-7. [DOI] [PubMed] [Google Scholar]
- Taylor J. W., Ott J., Eckstein F. The rapid generation of oligonucleotide-directed mutations at high frequency using phosphorothioate-modified DNA. Nucleic Acids Res. 1985 Dec 20;13(24):8765–8785. doi: 10.1093/nar/13.24.8765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsung K., Brissette R. E., Inouye M. Identification of the DNA-binding domain of the OmpR protein required for transcriptional activation of the ompF and ompC genes of Escherichia coli by in vivo DNA footprinting. J Biol Chem. 1989 Jun 15;264(17):10104–10109. [PubMed] [Google Scholar]
- Ullmann A. One-step purification of hybrid proteins which have beta-galactosidase activity. Gene. 1984 Jul-Aug;29(1-2):27–31. doi: 10.1016/0378-1119(84)90162-8. [DOI] [PubMed] [Google Scholar]
- Winans S. C., Allenza P., Stachel S. E., McBride K. E., Nester E. W. Characterization of the virE operon of the Agrobacterium Ti plasmid pTiA6. Nucleic Acids Res. 1987 Jan 26;15(2):825–837. doi: 10.1093/nar/15.2.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winans S. C., Ebert P. R., Stachel S. E., Gordon M. P., Nester E. W. A gene essential for Agrobacterium virulence is homologous to a family of positive regulatory loci. Proc Natl Acad Sci U S A. 1986 Nov;83(21):8278–8282. doi: 10.1073/pnas.83.21.8278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winans S. C., Kerstetter R. A., Nester E. W. Transcriptional regulation of the virA and virG genes of Agrobacterium tumefaciens. J Bacteriol. 1988 Sep;170(9):4047–4054. doi: 10.1128/jb.170.9.4047-4054.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winans S. C., Kerstetter R. A., Ward J. E., Nester E. W. A protein required for transcriptional regulation of Agrobacterium virulence genes spans the cytoplasmic membrane. J Bacteriol. 1989 Mar;171(3):1616–1622. doi: 10.1128/jb.171.3.1616-1622.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky M. F., Porter S. G., Young C., Albright L. M., Gordon M. P., Nester E. W. The virD operon of Agrobacterium tumefaciens encodes a site-specific endonuclease. Cell. 1986 Nov 7;47(3):471–477. doi: 10.1016/0092-8674(86)90604-5. [DOI] [PubMed] [Google Scholar]
- Zambryski P., Tempe J., Schell J. Transfer and function of T-DNA genes from agrobacterium Ti and Ri plasmids in plants. Cell. 1989 Jan 27;56(2):193–201. doi: 10.1016/0092-8674(89)90892-1. [DOI] [PubMed] [Google Scholar]