Abstract
Enzymes from thermophilic and hyperthermophilic organisms are invaluable catalysts for organic synthesis and biotechnology due to their stability and function at elevated temperatures. While the temperature dependency of catalysis with normal substrates has been studied for several of these enzymes, little is known about the effects of temperature on substrate specificity. Here we report the characterization of a novel thymidine kinase from the hyperthermophilic Thermotoga maritima which shows high substrate specificity at the organism's native growth temperature (82 °C) but turns promiscuous at 37 °C. Our experiments link this temperature-dependent substrate promiscuity to conformational changes in the protein below 70 °C, shifting the enzyme's rate-determining step to a substrate-independent transition in the homotetramer structure.
Enzymes from extremophilic organisms offer many interesting opportunities for applications in organic synthesis and biotechnology.1,2 They often exhibit tolerance for denaturants and organic solvents, as well as operate at extreme temperatures, pH and pressures. Enzymes from thermophilic and hyperthermophilic organisms are particularly attractive as increased stability and function at elevated temperature are desirable features for biocatalysis.
The temperature dependency of enzyme catalysis has been studied for several thermophilic biocatalysts, typically showing declining activity with decreasing temperature.3,4 This effect has been attributed to increasing rigidity of proteins at lower temperature. In contrast, little is known about how temperature might affect substrate specificity in enzyme catalysis. As part of our investigation of deoxynucleoside kinases (dNKs) for the phosphorylation of nucleoside analog (NA) prodrugs, we have isolated a novel thymidine kinase from the hyperthermophilic eubacterium Thermotoga maritima (TmTK) and explored the effects of temperature on the substrate specificity towards natural and unnatural nucleosides.
The gene for TmTK was isolated from T. maritima genomic DNA and heterologously overexpressed in E. coli. Following purification of the enzyme to homogeneity, crystallography confirmed its high similarity to human TK1, both in tertiary and quaternary structure.5 We are now reporting the catalytic performance of TmTK with various phosphoryl donors, as well as natural 2′-deoxyribonucleosides and NAs as phosphoryl acceptors at 37 °C and 82 °C, T. maritima's normal growth temperature (Table 1&S1). Initial kinetic measurements at 37°C indicate low donor specificity with a preference for ATP. In contrast, the enzyme is highly specific in regards to the nucleobase of the phosphoryl acceptor. Consistent with other members of the TK1-like family, TmTK phosphorylates only thymine and uracil-carrying substrates. The strict preference results from specific hydrogen-bonding interactions between the pyrimidine moiety of the substrate and the enzyme's lasso region.5,6
Table 1.
Kinetic parameters for TmTK substrates at 37°C & 82°C.
| substrate | KM (μM) | kcat (s−1) | kcat/KM × 104 (M−1 s−1) | relative efficiency | Hill coef. |
|---|---|---|---|---|---|
| T (37°C) | 0.5 ± 0.2 | 0.3 ± 0.08 | 60 | 1 | 0.98 |
| T (82°C) | 1.0 ± 0.15 | 3.5 ± 0.3 | 350 | 5.8 | 1.1 |
|
| |||||
| ATP (37°C) | 40 ± 12 | 0.4 ± 0.04 | 1.0 | 1 | 1.13 |
| ATP (82°C) | 187 ± 21(K0.5) | 3.5 ± 0.3 | 1.8 | 1.8 | 2.4 |
|
| |||||
| AZT (37°C) | 0.65 ± 0.2 | 0.29 ± 0.03 | 45 | 1 | - |
| AZT (82°C) | 8.7 ± 3 | 1.6 ± 0.29 | 18 | 0.4 | - |
At 82 °C, TmTK's catalytic rates, apparent binding constants, and cooperativity data for T and ATP closely resemble its mesophilic counterparts at their respective optimal temperatures (Table 1). Contrary to many hyperthermophilic enzymes, TmTK maintains significant activity at suboptimal temperatures. At 37 °C, the kcat/KM values for T and ATP are reduced by only 5.8- and 1.8-fold respectively. We also noticed the loss of ATP cooperativity in the homotetramer. More interestingly, the kinetic measurements with AZT at both temperatures showed the opposite trend. While AZT is phosphorylated with 20-fold lower efficiency than T at 82 °C, its kcat/KM value at 37 °C matches the natural substrate. More so, the enzyme is a more efficient catalyst for AZT phosphorylation at 37 °C than 82 °C, showing a 2.5-fold improvement in kcat/KM at the lower temperature. Hence, it appears that the enzyme can discriminate against unnatural substrates under physiological conditions at 82 °C, yet shows broader substrate tolerance at 37°C. Such temperature-dependent substrate promiscuity of extremozymes has not been reported previously but could be of fundamental interest in biocatalysis, enzyme engineering, and protein evolution.
To further investigate this phenomenon, we expanded our study to other NAs. While experiments at elevated temperatures were limited by the availability of tritiated substrates with sufficient specific activity, kinetic measurements with unlabeled NAs using a spectrophotometric coupled-enzyme assay at 37 °C confirmed the broad tolerance of TmTK for nucleosides with modified ribose moieties (Table 2). Differences in catalytic performance arise from variation in the KM values and correlate well with the preferred sugar conformation of the substrate. Crystallography and NMR spectroscopy, as well as studies with nucleosides in locked conformations have established the preferred C2′-endo sugar puckering of nucleoside bound to dNKs.7-9 TmTK follows the same trend, favoring substrates with C2′-endo and C3′-exo sugar puckering over ribose with a C3′-endo conformation. In addition, unfavorable steric effects are likely responsible for the decreased binding affinity of NAs carrying the L-isomer of 2′-deoxyribose or a ribose moiety. Similarly, KM values for 2′,3′-dideoxy-nucleosides increase due to missing stabilizing hydrogen-bonding interactions. In summary, the sugar puckering accounts for the differences in KM values at 37°C, yet does not explain TmTK's broadened substrate specificity compared to human TK1.
Table 2.
Kinetic parameters for nucleoside analogs at 37°C
| substrate | KM (μM) | kcat (s−1) | kcat/KM × 104 (M−1 s−1) | relative activity | preferred conform. |
|---|---|---|---|---|---|
| dU | 1.5 ± 1 | 0.7 ± 0.1 | 48 | 0.8 | C2′ -endo |
| U | > 1000 | n.d. | - | - | C3′ -endo |
| 5-FdUrd | 1 ± 0.1 | 0.74 ± 0.05 | 74 | 2.5 | C2′ -endo |
| ddT | 30 ± 5 | 0.33 ± 0.04 | 1.1 | 1.1 | C2′ -endo |
| ddU | > 1000 | n.d. | - | - | C3′ -endo |
| AZT | 0.65 ± 0.2 | 0.29 ± 0.03 | 45 | 1 | C3′ -endo |
| d4T | 75 ± 5 | 0.41 ± 0.02 | 0.54 | 1.4 | planar |
| DOT | 114 ± 15 | 0.29 ± 0.02 | 0.25 | 1 | C3′ -endo |
| L-FMAU | 132 ± 4 | 0.5 ± 0.02 | 0.38 | 1.7 | C2′ -endo |
| L-T | > 1000 | n.d. | - | - | C2′ -endo |
n.d. = not determined as vmax was not reached at 1000 μM substrate. dU: 2′-deoxyuridine, U: Uridine, ddT: 2′3′-dideoxythymidine, ddU: 2′3′-dideoxyuridine, 5FdUrd: 5-fluoro-2′-deoxyuridine, AZT: 2′,3′-dideoxy-3′-azidothymidine, d4T: 2′,3′-dideoxy-2′,3′-didehydrothymidine, L-FMAU: 2′-fluoro-5-methyl-β-L-arabinofuranosyluracil, L-T: L-thymidine, DOT: dioxolane thymidine
A more promising lead to explain the specificity change might be TmTK's strikingly similar catalytic rates for all tested nucleosides at 37 °C (Table 2). Mesophilic TK activity for the same substrates differ by up to two orders of magnitude.10 The rate homogeneity has been rationalized by a common intermediate preceding the rate-limiting step in the reaction pathway.11,12 While TmTK's direct phosphoryl-transfer mechanism makes a chemical intermediate implausible, the uniform kcat values are consistent with a common structural intermediate, resulting from a substrate-independent conformational change as the rate-determining step at 37 °C.
Evidence supporting such a structural change was found upon analysis of enzyme reaction rates at different temperature via Arrhenius plot (Figure 1). The data revealed a discontinuity at 70°C, indicating a change in the rate-limiting step. Above the transition temperature, the slope corresponds to an activation energy of ∼ 4 kcal/mol while below 70 °C, the energy barrier increases to 16 kcal/mol. A conformational change as the enzyme's slow-step at temperatures below 70 °C could explain the equal reaction rates.
Figure 1.
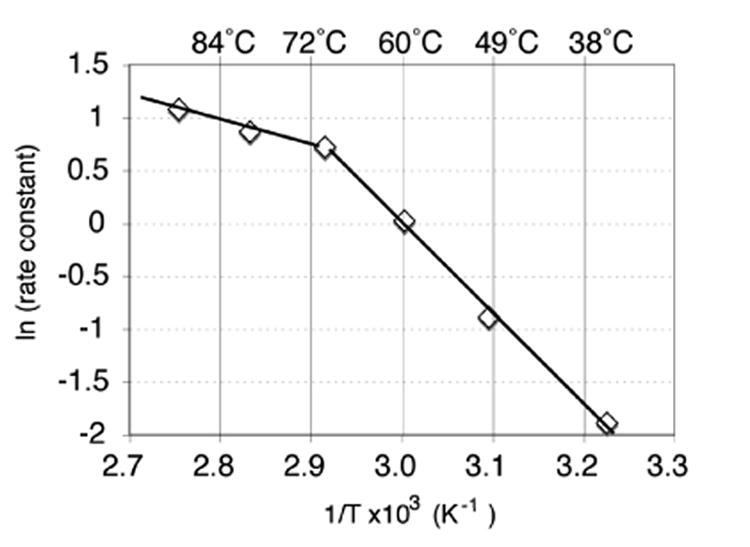
Arrhenius plot for TmTK with thymidine and ATP. The vmax values were measured by radioassay at the specified temperatures.
Independent verification for a temperature-dependent conformational change in TmTK was obtained by fluorescence spectroscopy. The crystal structure of the TmTK homotetramer suggests a transition at the dimer II interface upon ATP binding, shifting the equilibrium from a closed, inactive state to a catalytically competent, open conformation.5,13 Speculating that this transition could take place spontaneously at elevated temperature and correlates with our observed kinetic effect, we used mant-ADP, an ADP analog with an environmentally sensitive fluorescent N-methyl-anthraniloyl portion at the 2′ or 3′ position of the adenosine, to monitor conformational changes at the dimer II interface.
Our experiments detected a transition point in the fluorophore's emission wavelength at 70 °C (Figure 2). Monitoring the fluorescence intensity and λem of mant-ADP in the presence of T, we detected a biphasic pattern where λem showed minimal temperature dependence below 70 °C, yet blue shifted by 0.5 – 0.8 nm/°C above the transition temperature. While initial scans covered a temperature range from 37 °C to 90 °C, subsequent measurements in one-degree increments focused on a temperature range from 65 °C to 75 °C. As T/mant-ADP represents a mixed substrate/product complex, we repeated the experiments with TMP/mant-ADP and observed the same λem transition above 70 °C although with a more moderate slope of 0.2 nm/°C (data not shown). Control experiments with mant-ADP in the absence of TmTK show no significant temperature dependence of λem. The observed blue shift indicates a repositioning of the fluorophor in a more hydrophobic environment above 70 °C, consistent with the expansion at the dimer II interface.5
Figure 2.
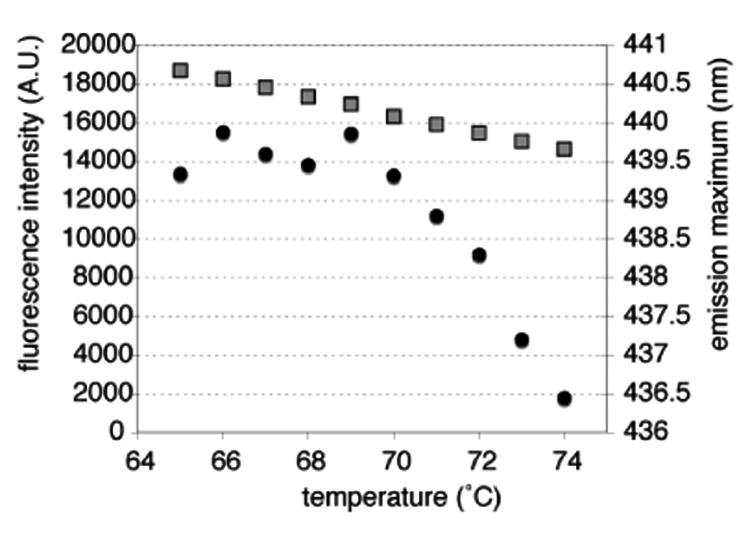
Fluorescence spectroscopy to detect temperature-dependent structural changes in TmTK via mant-ADP. The fluorescence intensity (□) and maximum emission wavelength (●) were plotted against temperature
In summary, our study of the TK from the T. maritima revealed a significant, temperature-dependent change in substrate specificity. While preferring thymidine to AZT at physiological conditions (82 °C), the enzyme shows substrate promiscuity at 37 °C. We have used kinetic and spectroscopic evidence to link this phenomenon to a conformational change in TmTK at 70 °C. Concurrent with structure data, we propose a two-state model for TmTK involving a thermodynamically preferred but inactive closed state and a catalytically competent open structure of the TmTK homotetramer. While in rapid equilibrium above 70 °C, the transition from closed to open state becomes the rate-limiting step of the reaction pathway at lower temperatures. Consequently, that leads to a normalization of the catalytic rates of phosphorylation and the observed substrate promiscuity of TmTK.
With respect to enzyme evolution and biocatalysis, future experiments will explore the generality of substrate promiscuity among enzymes from extremophiles and investigate the structural and functional consequences of changing protein dynamics in detail.
Supplementary Material
Acknowledgments
We acknowledge financial support in part by the NIH (GM69958) and by a grant to the Emory Center for AIDS Research (AI050409) from the NIH and by institutional funding from the Emory University HSC. We thank Dr. Raymond Schinazi for providing samples of the L-nucleosides and DOT.
Footnotes
Supporting Information Available: Full experimental details (PDF). This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Atomi H. Curr Opin Chem Biol. 2005;9:166–173. doi: 10.1016/j.cbpa.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Egorova K, Antranikian G. Curr Opin Microbiol. 2005;8:649–655. doi: 10.1016/j.mib.2005.10.015. [DOI] [PubMed] [Google Scholar]
- 3.Sehgal AC, Kelly RM. J Am Chem Soc. 2002;124:8190–8191. doi: 10.1021/ja026512q. [DOI] [PubMed] [Google Scholar]
- 4.Zavodszky P, Kardos J, Svingor A, Petsko GA. Proc Natl Acad Sci U S A. 1998;95:7406–7411. doi: 10.1073/pnas.95.13.7406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Segura-Pena D, Lutz S, Monnerjahn C, Konrad M, Lavie A. J Mol Biol. 2007;369:129–141. doi: 10.1016/j.jmb.2007.02.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Welin M, Kosinska U, Mikkelsen NE, Carnrot C, Zhu C, Wang L, Eriksson S, Munch-Petersen B, Eklund H. Proc Natl Acad Sci U S A. 2004;101:17970–17975. doi: 10.1073/pnas.0406332102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maltseva T, Usova E, Eriksson S, Milecki J, Foldesi A, Chattopadhayaya J. Nucl Nucl Nucl Acids. Vol. 20. 2001. pp. 1225–1228. [DOI] [PubMed] [Google Scholar]
- 8.Marquez VE, Ben-Kasus T, Barchi JJ, Green KM, Nicklaus MC, Agbaria R. J Am Chem Soc. 2004;126:543–549. doi: 10.1021/ja037929e. [DOI] [PubMed] [Google Scholar]
- 9.Sabini E, Ort S, Monnerjahn C, Konrad M, Lavie A. Nat Struct Biol. 2003;10:513–519. doi: 10.1038/nsb942. [DOI] [PubMed] [Google Scholar]
- 10.Eriksson S, Kierdaszuk B, Munch-Petersen B, Oberg B, Johansson NG. Biochem Biophys Res Commun. 1991;176:586–592. doi: 10.1016/s0006-291x(05)80224-4. [DOI] [PubMed] [Google Scholar]
- 11.Hollaway MR, Hardman MJ. Eur J Biochem. 1973;32:537–546. doi: 10.1111/j.1432-1033.1973.tb02639.x. [DOI] [PubMed] [Google Scholar]
- 12.Kirsch JF, Katchalski E. Biochemistry. 1965;4:884–890. doi: 10.1021/bi00881a014. [DOI] [PubMed] [Google Scholar]
- 13.Kosinska U, Carnrot C, Sandrini MP, Clausen AR, Wang L, Piskur J, Eriksson S, Eklund H. FEBS J. 2007;274:727–737. doi: 10.1111/j.1742-4658.2006.05617.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


