Abstract
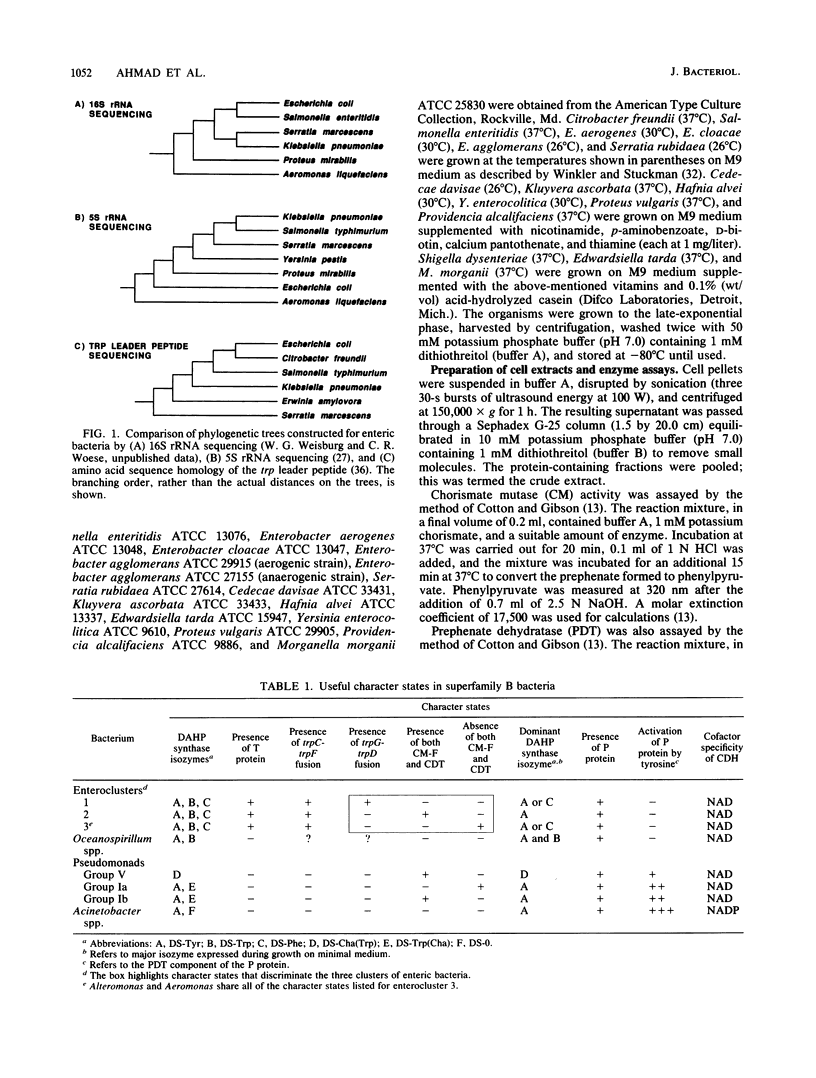
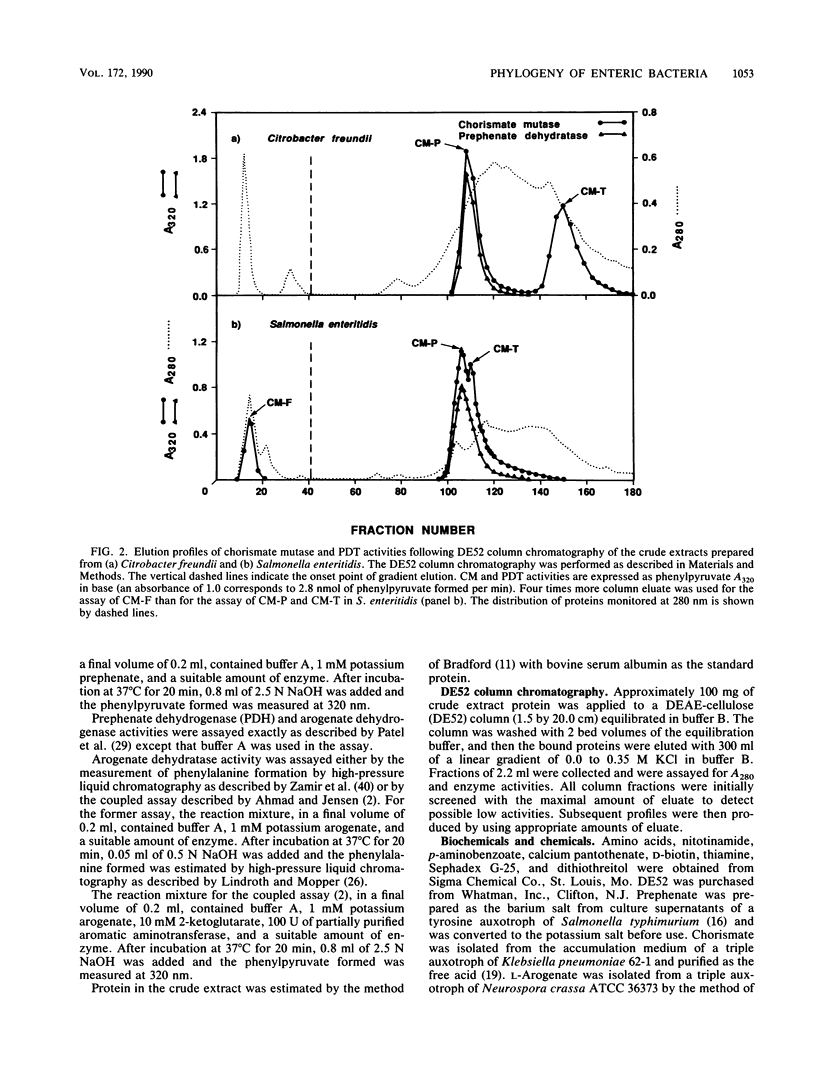
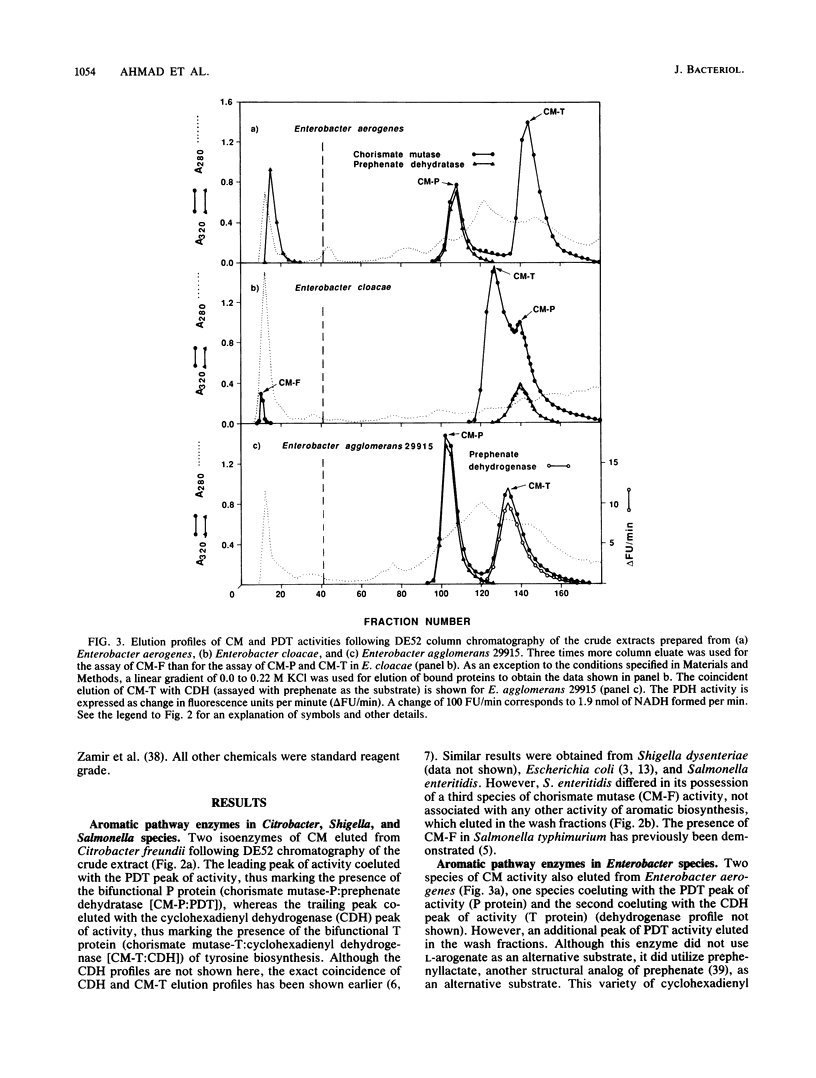
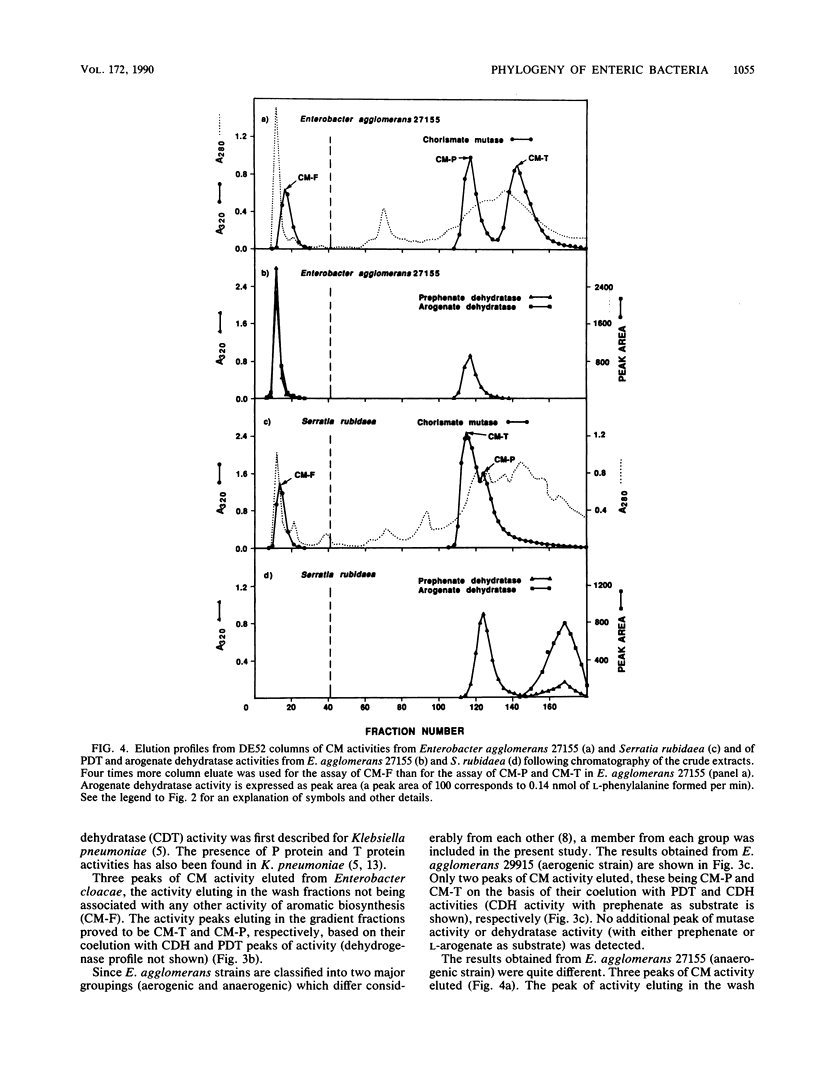
A comprehensive phylogenetic tree for virtually the entire assemblage of enteric bacteria is presented. Character states of aromatic amino acid biosynthesis are used as criteria, and the results are compared with partial trees based upon sequencing of 16S rRNA, 5S rRNA, and tryptophan leader peptide. Three major clusters are apparent. Enterocluster 1 possesses a gene fusion (trpG-trpD) encoding anthranilate synthase: anthranilate 5-phosphoribosylpyrophosphate phosphoribosyltransferase of tryptophan biosynthesis. This cluster includes the genera Escherichia, Shigella, Citrobacter, Salmonella, Klebsiella, and Enterobacter. The remaining two clusters lack the trpG-trpD gene fusion, but differ in the presence (enterocluster 2) or absence (enterocluster 3) of the three-step overflow pathway to L-phenylalanine. Enterocluster 2 consists of the genera Serratia and Erwinia. Enterocluster 3 includes the genera Cedecea, Kluyvera, Edwardsiella, Hafnia, Yersinia, Proteus, Providencia, and Morganella. Within these three major clusters, a tentative hierarchy of subcluster ordering is formulated on the basis of all data available. This hierarchical framework is proposed as a general working basis for continued refinement of the phylogenetic relationships of enteric bacteria.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ahmad S., Jensen R. A. A simple spectrophotometric assay for arogenate dehydratase. Anal Biochem. 1987 May 15;163(1):107–111. doi: 10.1016/0003-2697(87)90099-6. [DOI] [PubMed] [Google Scholar]
- Ahmad S., Jensen R. A. New prospects for deducing the evolutionary history of metabolic pathways in prokaryotes: aromatic biosynthesis as a case-in-point. Orig Life Evol Biosph. 1988;18(1-2):41–57. doi: 10.1007/BF01808779. [DOI] [PubMed] [Google Scholar]
- Ahmad S., Jensen R. A. The phylogenetic origin of the bifunctional tyrosine-pathway protein in the enteric lineage of bacteria. Mol Biol Evol. 1988 May;5(3):282–297. doi: 10.1093/oxfordjournals.molbev.a040496. [DOI] [PubMed] [Google Scholar]
- Ahmad S., Jensen R. A. The prephenate dehydrogenase component of the bifunctional T-protein in enteric bacteria can utilize L-arogenate. FEBS Lett. 1987 May 25;216(1):133–139. doi: 10.1016/0014-5793(87)80771-8. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Byng G. S., Kane J. F., Jensen R. A. Diversity in the routing and regulation of complex biochemical pathways as indicators of microbial relatedness. Crit Rev Microbiol. 1982 May;9(4):227–252. doi: 10.3109/10408418209104491. [DOI] [PubMed] [Google Scholar]
- COTTON R. G., GIBSON F. THE BIOSYNTHESIS OF PHENYLALANINE AND TYROSINE; ENZYMES CONVERTING CHORISMIC ACID INTO PREPHENIC ACID AND THEIR RELATIONSHIPS TO PREPHENATE DEHYDRATASE AND PREPHENATE DEHYDROGENASE. Biochim Biophys Acta. 1965 Apr 12;100:76–88. doi: 10.1016/0304-4165(65)90429-0. [DOI] [PubMed] [Google Scholar]
- Fiske M. J., Whitaker R. J., Jensen R. A. Hidden overflow pathway to L-phenylalanine in Pseudomonas aeruginosa. J Bacteriol. 1983 May;154(2):623–631. doi: 10.1128/jb.154.2.623-631.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox G. E., Stackebrandt E., Hespell R. B., Gibson J., Maniloff J., Dyer T. A., Wolfe R. S., Balch W. E., Tanner R. S., Magrum L. J. The phylogeny of prokaryotes. Science. 1980 Jul 25;209(4455):457–463. doi: 10.1126/science.6771870. [DOI] [PubMed] [Google Scholar]
- Harms E., Hsu J. H., Subrahmanyam C. S., Umbarger H. E. Comparison of the regulatory regions of ilvGEDA operons from several enteric organisms. J Bacteriol. 1985 Oct;164(1):207–216. doi: 10.1128/jb.164.1.207-216.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori H., Osawa S. Evolutionary change in 5S RNA secondary structure and a phylogenic tree of 54 5S RNA species. Proc Natl Acad Sci U S A. 1979 Jan;76(1):381–385. doi: 10.1073/pnas.76.1.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson G. S., Davidson B. E. Nucleotide sequence and transcription of the phenylalanine and tyrosine operons of Escherichia coli K12. J Mol Biol. 1984 Dec 25;180(4):1023–1051. doi: 10.1016/0022-2836(84)90269-9. [DOI] [PubMed] [Google Scholar]
- Jensen R. A. Biochemical pathways in prokaryotes can be traced backward through evolutionary time. Mol Biol Evol. 1985 Mar;2(2):92–108. doi: 10.1093/oxfordjournals.molbev.a040338. [DOI] [PubMed] [Google Scholar]
- Jensen R. A., Nasser D. S. Comparative regulation of isoenzymic 3-deoxy-D-arabino-heptulosonate 7-phosphate synthetases in microorganisms. J Bacteriol. 1968 Jan;95(1):188–196. doi: 10.1128/jb.95.1.188-196.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keener S. L., McNamee K. P., McEntee K. Cloning and characterization of recA genes froM Proteus vulgaris, Erwinia carotovora, Shigella flexneri, and Escherichia coli B/r. J Bacteriol. 1984 Oct;160(1):153–160. doi: 10.1128/jb.160.1.153-160.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonell M. T., Swartz D. G., Ortiz-Conde B. A., Last G. A., Colwell R. R. Ribosomal RNA phylogenies for the vibrio-enteric group of eubacteria. Microbiol Sci. 1986 Jun;3(6):172-5, 178. [PubMed] [Google Scholar]
- Nichols B. P., Miozzari G. F., van Cleemput M., Bennett G. N., Yanofsky C. Nucleotide sequences of the trpG regions of Escherichia coli, Shigella dysenteriae, Salmonella typhimurium and Serratia marcescens. J Mol Biol. 1980 Oct 5;142(4):503–517. doi: 10.1016/0022-2836(80)90260-0. [DOI] [PubMed] [Google Scholar]
- Patel N., Pierson D. L., Jensen R. A. Dual enzymatic routes to L-tyrosine and L-phenylalanine via pretyrosine in Pseudomonas aeruginosa. J Biol Chem. 1977 Aug 25;252(16):5839–5846. [PubMed] [Google Scholar]
- Shiuan D., Campbell A. Transcriptional regulation and gene arrangement of Escherichia coli, Citrobacter freundii and Salmonella typhimurium biotin operons. Gene. 1988 Jul 30;67(2):203–211. doi: 10.1016/0378-1119(88)90397-6. [DOI] [PubMed] [Google Scholar]
- Winkler U. K., Stuckmann M. Glycogen, hyaluronate, and some other polysaccharides greatly enhance the formation of exolipase by Serratia marcescens. J Bacteriol. 1979 Jun;138(3):663–670. doi: 10.1128/jb.138.3.663-670.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R. Bacterial evolution. Microbiol Rev. 1987 Jun;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata H., Nakamura K., Inouye M. Comparison of the lipoprotein gene among the Enterobacteriaceae. DNA sequence of Erwinia amylovora lipoprotein gene. J Biol Chem. 1981 Mar 10;256(5):2194–2198. [PubMed] [Google Scholar]
- Yanofsky C. Comparison of regulatory and structural regions of genes of tryptophan metabolism. Mol Biol Evol. 1984 Feb;1(2):143–161. doi: 10.1093/oxfordjournals.molbev.a040307. [DOI] [PubMed] [Google Scholar]
- Zamir L. O., Tiberio R., Devor K. A., Sauriol F., Ahmad S., Jensen R. A. Structure of D-prephenyllactate. A carboxycyclohexadienyl metabolite from Neurospora crassa. J Biol Chem. 1988 Nov 25;263(33):17284–17290. [PubMed] [Google Scholar]
- Zamir L. O., Tiberio R., Fiske M., Berry A., Jensen R. A. Enzymatic and nonenzymatic dehydration reactions of L-arogenate. Biochemistry. 1985 Mar 26;24(7):1607–1612. doi: 10.1021/bi00328a006. [DOI] [PubMed] [Google Scholar]


