Abstract
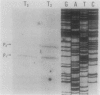
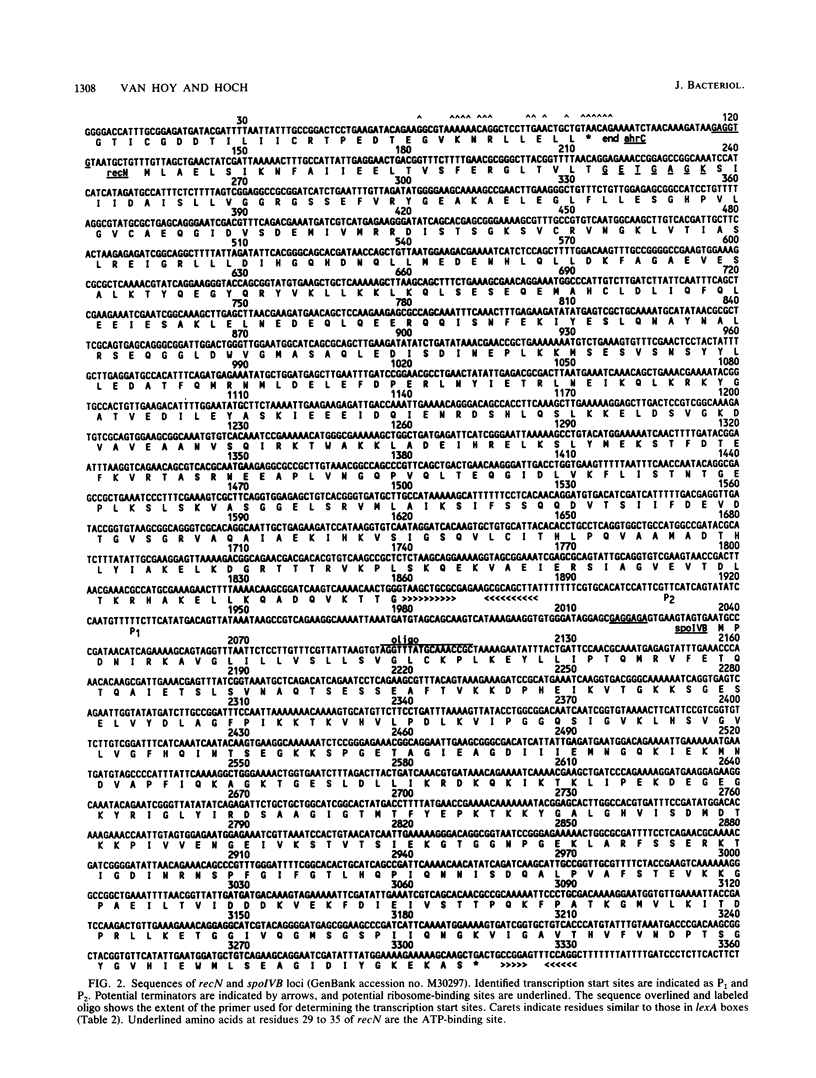
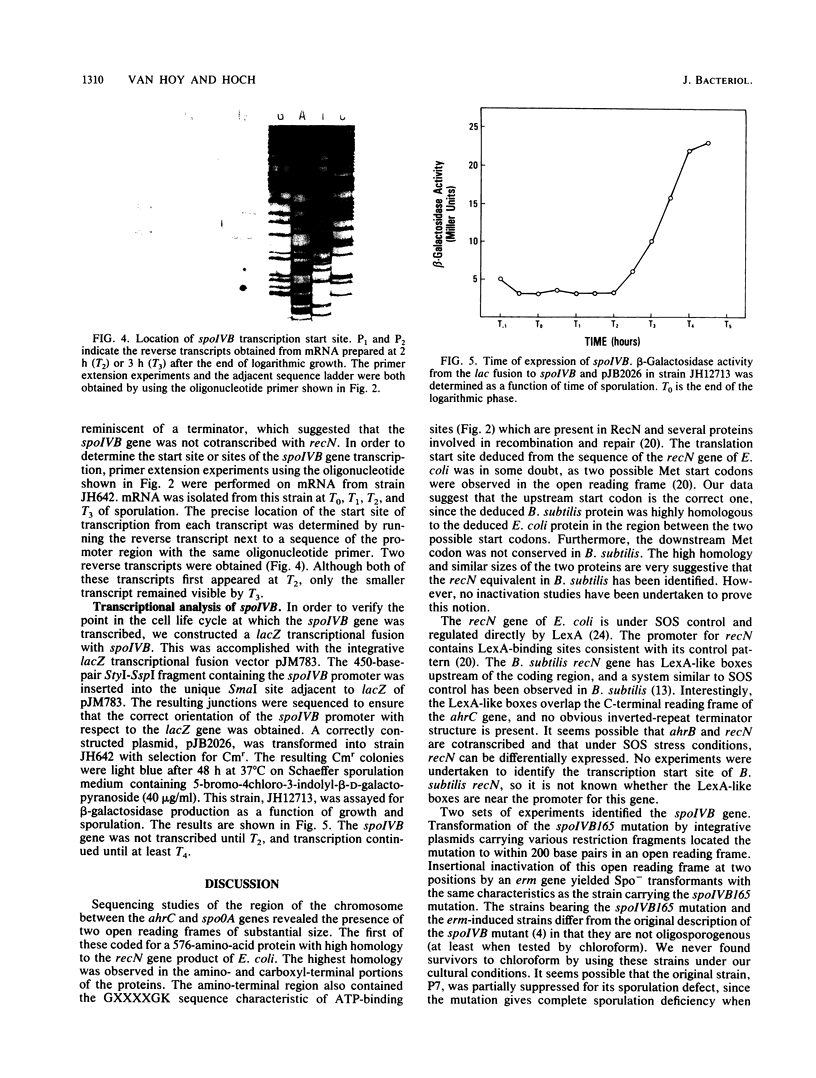
Two independent genes, recN and spoIVB, along with their respective promoter and termination regions, were discovered and sequenced in the 3.4-kilobase region between the ahrC and spoOA genes at map position 216 in the Bacillus subtilis chromosome map. The gene encoding a 576-amino-acid protein, which maintains a high homology with the Escherichia coli recN gene product, was adjacent to ahrC. The sequence revealed a 64,472-dalton polypeptide which contained a conserved ATP-binding site and possible lexA-type regulatory binding sequences in its promoter region. A second open reading frame identified as the spoIVB gene was directly downstream of recN. It consisted of 1,275 nucleotides which coded for a 425-amino-acid polypeptide with a molecular weight of 45,976. Phenotypic, genetic, and transcriptional analyses confirmed that this gene was spoIVB. Although no chloroform-resistant spores were produced by spoIVB-inactivated strains, under microscopic examination, phase-gray forespores were visible. The spoIVB165 mutation was localized to a 200-base-pair region in the amino-terminal portion of the polypeptide, spoIVB was not transcribed until hour 2 of sporulation in wild-type B. subtilis cells, as determined by beta-galactosidase activity assays from lacZ transcriptional fusion constructions. We found no amino acid sequence homology between the spoIVB gene product and other known bacterial proteins.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anagnostopoulos C., Spizizen J. REQUIREMENTS FOR TRANSFORMATION IN BACILLUS SUBTILIS. J Bacteriol. 1961 May;81(5):741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E. Y., Seeburg P. H. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985 Apr;4(2):165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- Coote J. G. Sporulation in Bacillus subtilis. Characterization of oligosporogenous mutants and comparison of their phenotypes with those of asporogenous mutants. J Gen Microbiol. 1972 Jun;71(1):1–15. doi: 10.1099/00221287-71-1-1. [DOI] [PubMed] [Google Scholar]
- Coote J. G. Sporulation in Bacillus subtilis. Genetic analysis of oligosporogenous mutants. J Gen Microbiol. 1972 Jun;71(1):17–27. doi: 10.1099/00221287-71-1-17. [DOI] [PubMed] [Google Scholar]
- Ferrari E., Henner D. J., Perego M., Hoch J. A. Transcription of Bacillus subtilis subtilisin and expression of subtilisin in sporulation mutants. J Bacteriol. 1988 Jan;170(1):289–295. doi: 10.1128/jb.170.1.289-295.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari F. A., Trach K., LeCoq D., Spence J., Ferrari E., Hoch J. A. Characterization of the spo0A locus and its deduced product. Proc Natl Acad Sci U S A. 1985 May;82(9):2647–2651. doi: 10.1073/pnas.82.9.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins D. G., Sharp P. M. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene. 1988 Dec 15;73(1):237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- Hoch J. A. Genetics of bacterial sporulation. Adv Genet. 1976;18:69–98. doi: 10.1016/s0065-2660(08)60437-x. [DOI] [PubMed] [Google Scholar]
- Hoch J. A. Selection of cells transformed to prototrophy for sporulation markers. J Bacteriol. 1971 Mar;105(3):1200–1201. doi: 10.1128/jb.105.3.1200-1201.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes D. S., Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981 Jun;114(1):193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- Losick R., Youngman P., Piggot P. J. Genetics of endospore formation in Bacillus subtilis. Annu Rev Genet. 1986;20:625–669. doi: 10.1146/annurev.ge.20.120186.003205. [DOI] [PubMed] [Google Scholar]
- Love P. E., Yasbin R. E. Genetic characterization of the inducible SOS-like system of Bacillus subtilis. J Bacteriol. 1984 Dec;160(3):910–920. doi: 10.1128/jb.160.3.910-920.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninfa A. J., Magasanik B. Covalent modification of the glnG product, NRI, by the glnL product, NRII, regulates the transcription of the glnALG operon in Escherichia coli. Proc Natl Acad Sci U S A. 1986 Aug;83(16):5909–5913. doi: 10.1073/pnas.83.16.5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North A. K., Smith M. C., Baumberg S. Nucleotide sequence of a Bacillus subtilis arginine regulatory gene and homology of its product to the Escherichia coli arginine repressor. Gene. 1989 Aug 1;80(1):29–38. doi: 10.1016/0378-1119(89)90247-3. [DOI] [PubMed] [Google Scholar]
- Perego M., Spiegelman G. B., Hoch J. A. Structure of the gene for the transition state regulator, abrB: regulator synthesis is controlled by the spo0A sporulation gene in Bacillus subtilis. Mol Microbiol. 1988 Nov;2(6):689–699. doi: 10.1111/j.1365-2958.1988.tb00079.x. [DOI] [PubMed] [Google Scholar]
- Piggot P. J., Coote J. G. Genetic aspects of bacterial endospore formation. Bacteriol Rev. 1976 Dec;40(4):908–962. doi: 10.1128/br.40.4.908-962.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronson C. W., Nixon B. T., Ausubel F. M. Conserved domains in bacterial regulatory proteins that respond to environmental stimuli. Cell. 1987 Jun 5;49(5):579–581. doi: 10.1016/0092-8674(87)90530-7. [DOI] [PubMed] [Google Scholar]
- Rostas K., Morton S. J., Picksley S. M., Lloyd R. G. Nucleotide sequence and LexA regulation of the Escherichia coli recN gene. Nucleic Acids Res. 1987 Jul 10;15(13):5041–5049. doi: 10.1093/nar/15.13.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer R. T., Yocum R. R., Doolittle R. F., Lewis M., Pabo C. O. Homology among DNA-binding proteins suggests use of a conserved super-secondary structure. Nature. 1982 Jul 29;298(5873):447–451. doi: 10.1038/298447a0. [DOI] [PubMed] [Google Scholar]
- Schaeffer P., Millet J., Aubert J. P. Catabolic repression of bacterial sporulation. Proc Natl Acad Sci U S A. 1965 Sep;54(3):704–711. doi: 10.1073/pnas.54.3.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trach K., Chapman J. W., Piggot P., LeCoq D., Hoch J. A. Complete sequence and transcriptional analysis of the spo0F region of the Bacillus subtilis chromosome. J Bacteriol. 1988 Sep;170(9):4194–4208. doi: 10.1128/jb.170.9.4194-4208.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker G. C. Mutagenesis and inducible responses to deoxyribonucleic acid damage in Escherichia coli. Microbiol Rev. 1984 Mar;48(1):60–93. doi: 10.1128/mr.48.1.60-93.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]