Abstract
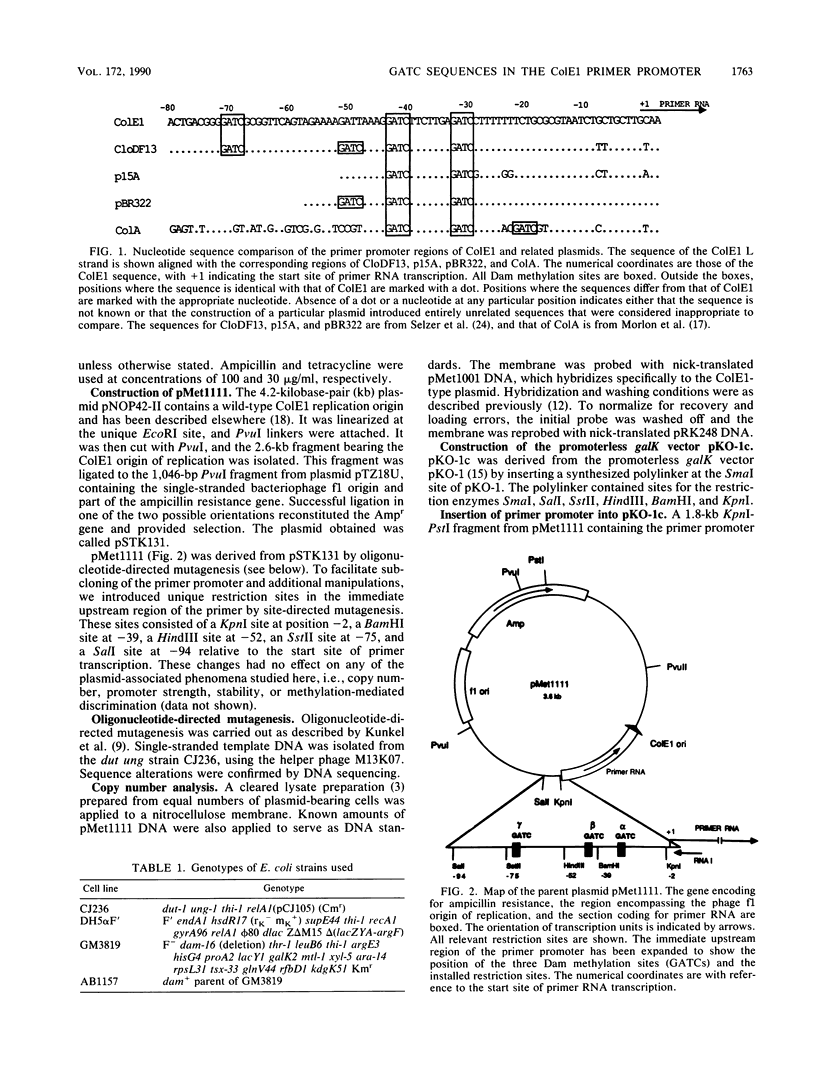
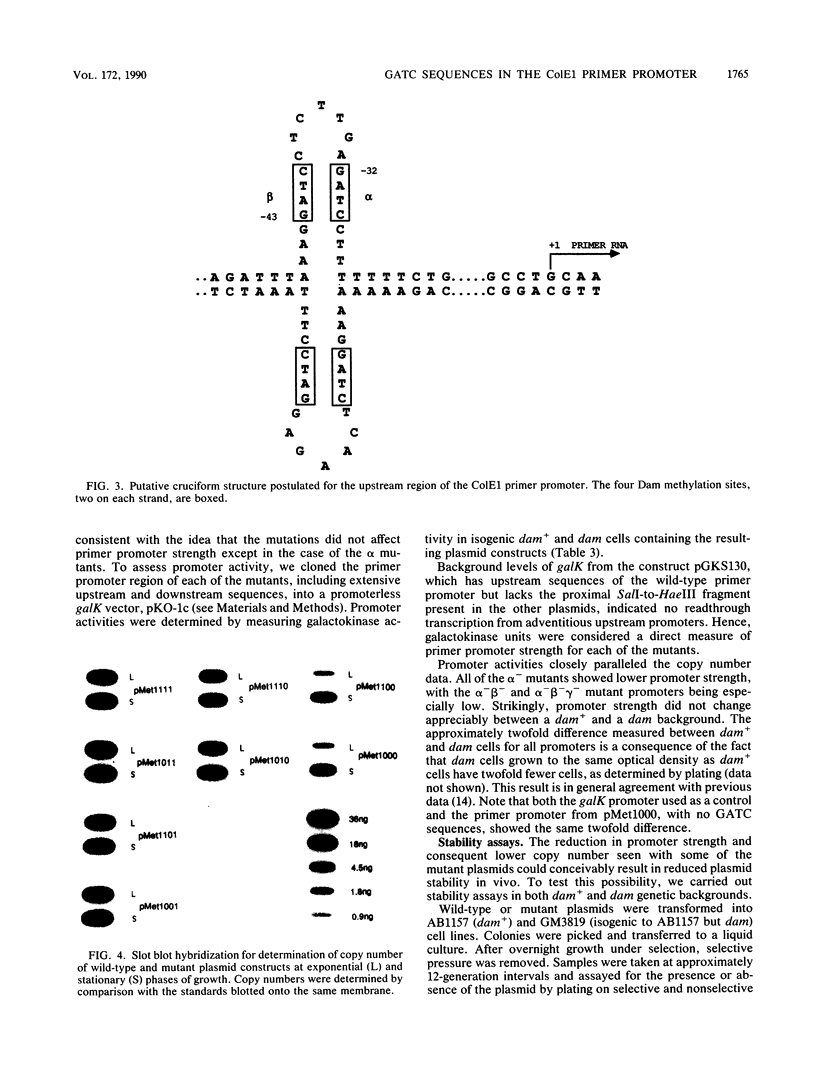
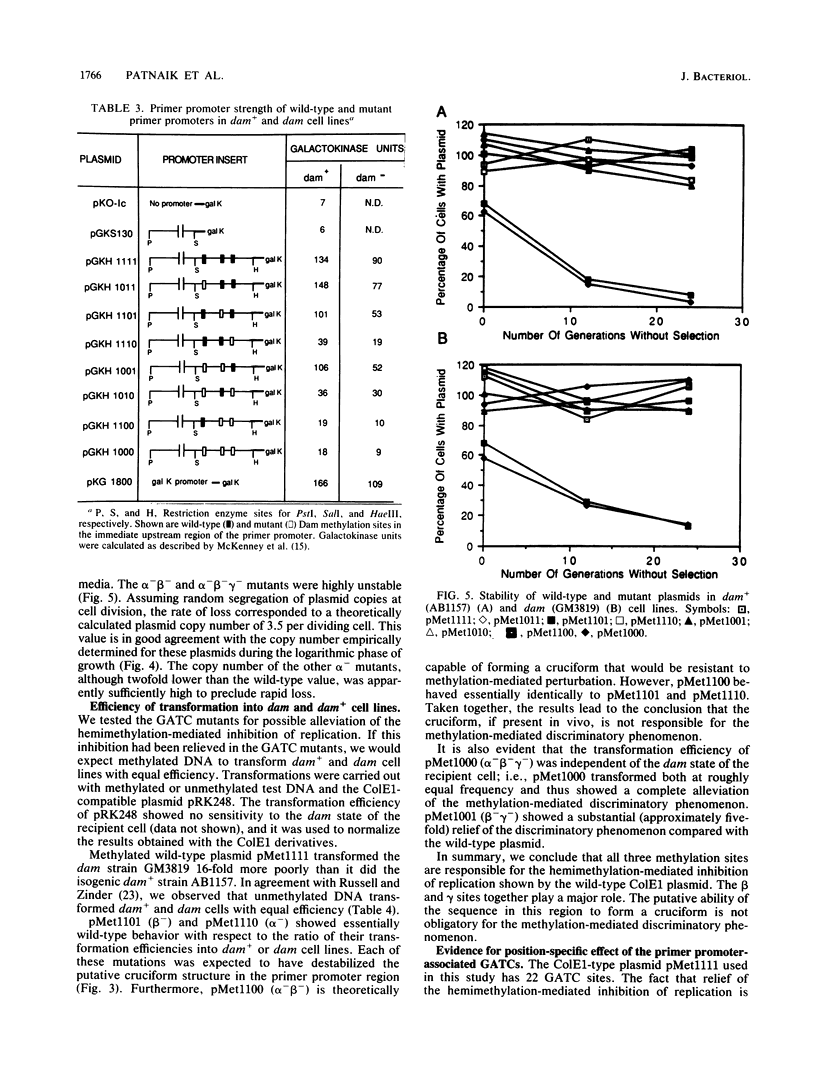
Plasmid ColE1 has three recognition sites for the Escherichia coli DNA adenine methylase in the immediate upstream region of the primer promoter. Two of these sites are conserved among all plasmid relatives of ColE1 and constitute parts of an inverted repeat that can conceivably form a cruciform structure. Recent experiments have indicated that hemimethylated ColE1-type plasmids are inefficiently replicated after transformation (D. W. Russell and N. Zinder, Cell 50:1071-1079, 1987). By mutating the three methylation sites, we found that disruption of all three GATC sites was necessary for complete relief of the hemimethylation-mediated inhibition of replication in vivo. We also found that these three methylation sites acted in a position-specific manner. The putative cruciform, if present, did not play a regulatory role in the hemimethylation-mediated inhibition of replication.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abeles A. L., Austin S. J. P1 plasmid replication requires methylated DNA. EMBO J. 1987 Oct;6(10):3185–3189. doi: 10.1002/j.1460-2075.1987.tb02630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barras F., Marinus M. G. The great GATC: DNA methylation in E. coli. Trends Genet. 1989 May;5(5):139–143. doi: 10.1016/0168-9525(89)90054-1. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun R. E., Wright A. DNA methylation differentially enhances the expression of one of the two E. coli dnaA promoters in vivo and in vitro. Mol Gen Genet. 1986 Feb;202(2):246–250. doi: 10.1007/BF00331644. [DOI] [PubMed] [Google Scholar]
- Bölker M., Kahmann R. The Escherichia coli regulatory protein OxyR discriminates between methylated and unmethylated states of the phage Mu mom promoter. EMBO J. 1989 Aug;8(8):2403–2410. doi: 10.1002/j.1460-2075.1989.tb08370.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan P. T., Ohmori H., Tomizawa J., Lebowitz J. Nucleotide sequence and gene organization of ColE1 DNA. J Biol Chem. 1985 Jul 25;260(15):8925–8935. [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Hsu L. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2110–2114. doi: 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson P., Wolf-Watz H., Lind L., Johansson K. E., Nordström K. Binding between the par region of plasmids R1 and pSC101 and the outer membrane fraction of the host bacteria. EMBO J. 1983;2(1):27–32. doi: 10.1002/j.1460-2075.1983.tb01375.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Leonard A. C., Helmstetter C. E. Cell cycle-specific replication of Escherichia coli minichromosomes. Proc Natl Acad Sci U S A. 1986 Jul;83(14):5101–5105. doi: 10.1073/pnas.83.14.5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel M., Higa A. Calcium-dependent bacteriophage DNA infection. J Mol Biol. 1970 Oct 14;53(1):159–162. doi: 10.1016/0022-2836(70)90051-3. [DOI] [PubMed] [Google Scholar]
- Marinus M. G. DNA methylation in Escherichia coli. Annu Rev Genet. 1987;21:113–131. doi: 10.1146/annurev.ge.21.120187.000553. [DOI] [PubMed] [Google Scholar]
- Marinus M. G., Morris N. R. Biological function for 6-methyladenine residues in the DNA of Escherichia coli K12. J Mol Biol. 1974 May 15;85(2):309–322. doi: 10.1016/0022-2836(74)90366-0. [DOI] [PubMed] [Google Scholar]
- Messer W., Noyer-Weidner M. Timing and targeting: the biological functions of Dam methylation in E. coli. Cell. 1988 Sep 9;54(6):735–737. doi: 10.1016/s0092-8674(88)90911-7. [DOI] [PubMed] [Google Scholar]
- Morlon J., Chartier M., Bidaud M., Lazdunski C. The complete nucleotide sequence of the colicinogenic plasmid ColA. High extent of homology with ColE1. Mol Gen Genet. 1988 Feb;211(2):231–243. doi: 10.1007/BF00330599. [DOI] [PubMed] [Google Scholar]
- Muesing M., Tamm J., Shepard H. M., Polisky B. A single base-pair alteration is responsible for the DNA overproduction phenotype of a plasmid copy-number mutant. Cell. 1981 Apr;24(1):235–242. doi: 10.1016/0092-8674(81)90519-5. [DOI] [PubMed] [Google Scholar]
- Murchie A. I., Lilley D. M. Base methylation and local DNA helix stability. Effect on the kinetics of cruciform extrusion. J Mol Biol. 1989 Feb 5;205(3):593–602. doi: 10.1016/0022-2836(89)90228-3. [DOI] [PubMed] [Google Scholar]
- Ogden G. B., Pratt M. J., Schaechter M. The replicative origin of the E. coli chromosome binds to cell membranes only when hemimethylated. Cell. 1988 Jul 1;54(1):127–135. doi: 10.1016/0092-8674(88)90186-9. [DOI] [PubMed] [Google Scholar]
- Polisky B. ColE1 replication control circuitry: sense from antisense. Cell. 1988 Dec 23;55(6):929–932. doi: 10.1016/0092-8674(88)90235-8. [DOI] [PubMed] [Google Scholar]
- Roberts D., Hoopes B. C., McClure W. R., Kleckner N. IS10 transposition is regulated by DNA adenine methylation. Cell. 1985 Nov;43(1):117–130. doi: 10.1016/0092-8674(85)90017-0. [DOI] [PubMed] [Google Scholar]
- Russell D. W., Zinder N. D. Hemimethylation prevents DNA replication in E. coli. Cell. 1987 Sep 25;50(7):1071–1079. doi: 10.1016/0092-8674(87)90173-5. [DOI] [PubMed] [Google Scholar]
- Selzer G., Som T., Itoh T., Tomizawa J. The origin of replication of plasmid p15A and comparative studies on the nucleotide sequences around the origin of related plasmids. Cell. 1983 Jan;32(1):119–129. doi: 10.1016/0092-8674(83)90502-0. [DOI] [PubMed] [Google Scholar]
- Smith D. W., Garland A. M., Herman G., Enns R. E., Baker T. A., Zyskind J. W. Importance of state of methylation of oriC GATC sites in initiation of DNA replication in Escherichia coli. EMBO J. 1985 May;4(5):1319–1326. doi: 10.1002/j.1460-2075.1985.tb03779.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomizawa J., Itoh T., Selzer G., Som T. Inhibition of ColE1 RNA primer formation by a plasmid-specified small RNA. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1421–1425. doi: 10.1073/pnas.78.3.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zyskind J. W., Cleary J. M., Brusilow W. S., Harding N. E., Smith D. W. Chromosomal replication origin from the marine bacterium Vibrio harveyi functions in Escherichia coli: oriC consensus sequence. Proc Natl Acad Sci U S A. 1983 Mar;80(5):1164–1168. doi: 10.1073/pnas.80.5.1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zyskind J. W., Smith D. W. The bacterial origin of replication, oriC. Cell. 1986 Aug 15;46(4):489–490. doi: 10.1016/0092-8674(86)90873-1. [DOI] [PubMed] [Google Scholar]
- van Putten A. J., de Lang R., Veltkamp E., Nijkamp H. J., Van Solingen P., van den Berg J. A. Methylation-dependent transcription controls plasmid replication of the CloDF13 cop-1(Ts) mutant. J Bacteriol. 1986 Nov;168(2):728–733. doi: 10.1128/jb.168.2.728-733.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]