Abstract
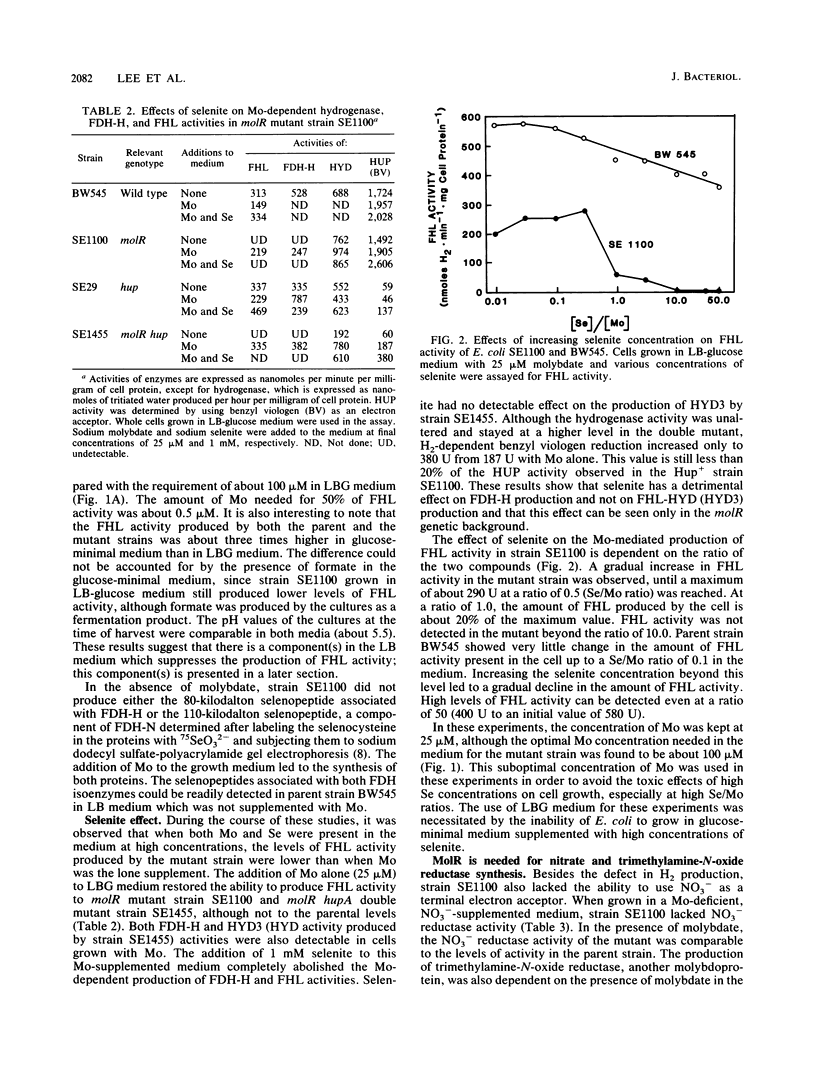
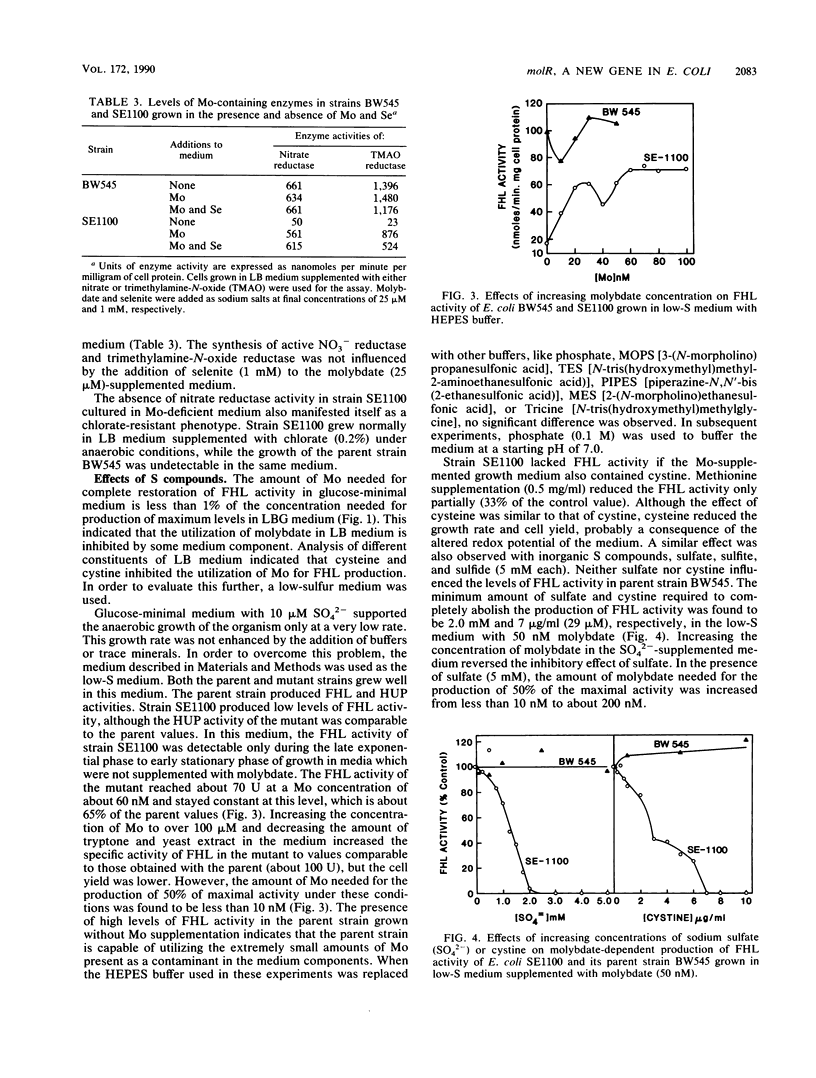
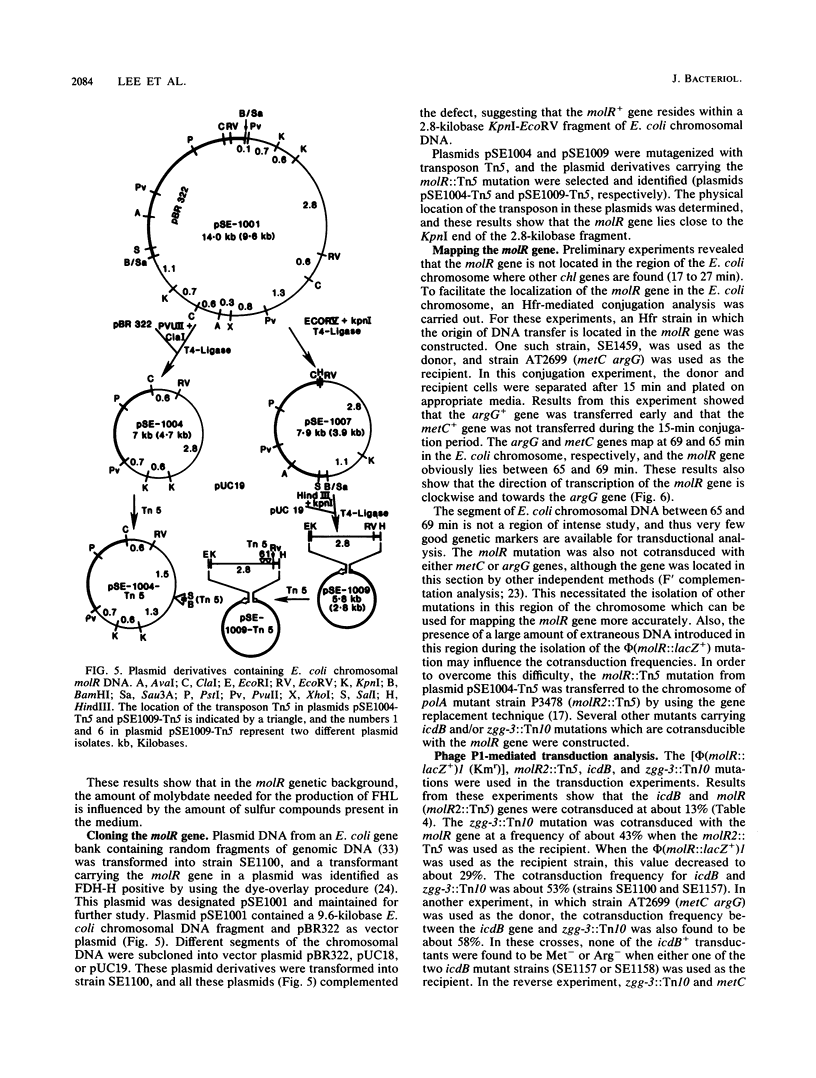
A mutation in a new gene, molR, prevented the synthesis in Escherichia coli of molybdoenzymes, including the two formate dehydrogenase isoenzymes, nitrate reductase and trimethylamine-N-oxide reductase. This phenotype was suppressed by supplementing the media with molybdate. Thus, the molR mutant was phenotypically similar to previously described chlD mutants, thought to be defective in molybdate transport. The molR gene is located at 65.3 min in the E. coli chromosome, in contrast to the chlD gene, which maps at 17 min and thus can be readily distinguished. The molR gene is also cotransducible with a hitherto unidentified gene essential for the production of 2-oxoglutarate from isocitrate, designated icdB (located at 66 min). The molR mutant strain SE1100 also failed to produce the hydrogenase component of formate hydrogenlyase (HYD3) in molybdate-unsupplemented media. The amount of molybdate required by strain SE1100 for the production of parental levels of formate hydrogenlyase activity was dependent on the growth medium. In Luria-Bertani medium, this value was about 100 microM, and in glucose-minimal medium, 1.0 microM was sufficient. In low-sulfur medium, this value decreased to about 50 nM. The addition of sulfate or selenite increased the amount of molybdate needed for the production of formate hydrogenlyase activity. These data suggest that in the absence of the high-affinity molybdate transport system, E. coli utilizes sulfate and selenite transport systems for transporting molybdate, preferring sulfate transport over the selenite transport system.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ames G. F. Bacterial periplasmic transport systems: structure, mechanism, and evolution. Annu Rev Biochem. 1986;55:397–425. doi: 10.1146/annurev.bi.55.070186.002145. [DOI] [PubMed] [Google Scholar]
- Bachmann B. J. Linkage map of Escherichia coli K-12, edition 7. Microbiol Rev. 1983 Jun;47(2):180–230. doi: 10.1128/mr.47.2.180-230.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballantine S. P., Boxer D. H. Isolation and characterisation of a soluble active fragment of hydrogenase isoenzyme 2 from the membranes of anaerobically grown Escherichia coli. Eur J Biochem. 1986 Apr 15;156(2):277–284. doi: 10.1111/j.1432-1033.1986.tb09578.x. [DOI] [PubMed] [Google Scholar]
- Ballantine S. P., Boxer D. H. Nickel-containing hydrogenase isoenzymes from anaerobically grown Escherichia coli K-12. J Bacteriol. 1985 Aug;163(2):454–459. doi: 10.1128/jb.163.2.454-459.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremer E., Silhavy T. J., Weisemann J. M., Weinstock G. M. Lambda placMu: a transposable derivative of bacteriophage lambda for creating lacZ protein fusions in a single step. J Bacteriol. 1984 Jun;158(3):1084–1093. doi: 10.1128/jb.158.3.1084-1093.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell A. M., del Campillo-Campbell A., Villaret D. B. Molybdate reduction by Escherichia coli K-12 and its chl mutants. Proc Natl Acad Sci U S A. 1985 Jan;82(1):227–231. doi: 10.1073/pnas.82.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chumley F. G., Menzel R., Roth J. R. Hfr formation directed by tn10. Genetics. 1979 Apr;91(4):639–655. doi: 10.1093/genetics/91.4.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J. C., Edwards E. S., DeMoss J. A. Resolution of distinct selenium-containing formate dehydrogenases from Escherichia coli. J Bacteriol. 1981 Mar;145(3):1317–1324. doi: 10.1128/jb.145.3.1317-1324.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DREYFUSS J. CHARACTERIZATION OF A SULFATE- AND THIOSULFATE-TRANSPORTING SYSTEM IN SALMONELLA TYPHIMURIUM. J Biol Chem. 1964 Jul;239:2292–2297. [PubMed] [Google Scholar]
- Dreyfuss J., Pardee A. B. Regulation of sulfate transport in Salmonella typhimurium. J Bacteriol. 1966 Jun;91(6):2275–2280. doi: 10.1128/jb.91.6.2275-2280.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott B. B., Mortenson L. E. Transport of molybdate by Clostridium pasteurianum. J Bacteriol. 1975 Dec;124(3):1295–1301. doi: 10.1128/jb.124.3.1295-1301.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster T. J., Lundblad V., Hanley-Way S., Halling S. M., Kleckner N. Three Tn10-associated excision events: relationship to transposition and role of direct and inverted repeats. Cell. 1981 Jan;23(1):215–227. doi: 10.1016/0092-8674(81)90286-5. [DOI] [PubMed] [Google Scholar]
- GRAY C. T., GEST H. BIOLOGICAL FORMATION OF MOLECULAR HYDROGEN. Science. 1965 Apr 9;148(3667):186–192. doi: 10.1126/science.148.3667.186. [DOI] [PubMed] [Google Scholar]
- Glaser J. H., DeMoss J. A. Phenotypic restoration by molybdate of nitrate reductase activity in chlD mutants of Escherichia coli. J Bacteriol. 1971 Nov;108(2):854–860. doi: 10.1128/jb.108.2.854-860.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutterson N. I., Koshland D. E., Jr Replacement and amplification of bacterial genes with sequences altered in vitro. Proc Natl Acad Sci U S A. 1983 Aug;80(16):4894–4898. doi: 10.1073/pnas.80.16.4894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johann S., Hinton S. M. Cloning and nucleotide sequence of the chlD locus. J Bacteriol. 1987 May;169(5):1911–1916. doi: 10.1128/jb.169.5.1911-1916.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J. L., Hainline B. E., Rajagopalan K. V., Arison B. H. The pterin component of the molybdenum cofactor. Structural characterization of two fluorescent derivatives. J Biol Chem. 1984 May 10;259(9):5414–5422. [PubMed] [Google Scholar]
- Lee J. H., Patel P., Sankar P., Shanmugam K. T. Isolation and characterization of mutant strains of Escherichia coli altered in H2 metabolism. J Bacteriol. 1985 Apr;162(1):344–352. doi: 10.1128/jb.162.1.344-352.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low K. B. Escherichia coli K-12 F-prime factors, old and new. Bacteriol Rev. 1972 Dec;36(4):587–607. doi: 10.1128/br.36.4.587-607.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. B., Scott D. J., Amy N. K. Molybdenum-sensitive transcriptional regulation of the chlD locus of Escherichia coli. J Bacteriol. 1987 May;169(5):1853–1860. doi: 10.1128/jb.169.5.1853-1860.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murgola E. J., Yanofsky C. Structural interactions between amino acid residues at positions 22 and 211 in the tryptophan synthetase alpha chain of Escherichia coli. J Bacteriol. 1974 Feb;117(2):444–448. doi: 10.1128/jb.117.2.444-448.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PASTERNAK C. A., ELLIS R. J., JONES-MORTIMER M. C., CRICHTON C. E. THE CONTROL OF SULPHATE REDUCTION IN BACTERIA. Biochem J. 1965 Jul;96:270–275. doi: 10.1042/bj0960270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PECK H. D., Jr, GEST H. Formic dehydrogenase and the hydrogenlyase enzyme complex in coli-aerogenes bacteria. J Bacteriol. 1957 Jun;73(6):706–721. doi: 10.1128/jb.73.6.706-721.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardee A. B., Prestidge L. S., Whipple M. B., Dreyfuss J. A binding site for sulfate and its relation to sulfate transport into Salmonella typhimurium. J Biol Chem. 1966 Sep 10;241(17):3962–3969. [PubMed] [Google Scholar]
- Pecher A., Zinoni F., Böck A. The seleno-polypeptide of formic dehydrogenase (formate hydrogen-lyase linked) from Escherichia coli: genetic analysis. Arch Microbiol. 1985 May;141(4):359–363. doi: 10.1007/BF00428850. [DOI] [PubMed] [Google Scholar]
- Sankar P., Lee J. H., Shanmugam K. T. Cloning of hydrogenase genes and fine structure analysis of an operon essential for H2 metabolism in Escherichia coli. J Bacteriol. 1985 Apr;162(1):353–360. doi: 10.1128/jb.162.1.353-360.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankar P., Shanmugam K. T. Biochemical and genetic analysis of hydrogen metabolism in Escherichia coli: the hydB gene. J Bacteriol. 1988 Dec;170(12):5433–5439. doi: 10.1128/jb.170.12.5433-5439.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawers R. G., Ballantine S. P., Boxer D. H. Differential expression of hydrogenase isoenzymes in Escherichia coli K-12: evidence for a third isoenzyme. J Bacteriol. 1985 Dec;164(3):1324–1331. doi: 10.1128/jb.164.3.1324-1331.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlensog V., Birkmann A., Böck A. Mutations in trans which affect the anaerobic expression of a formate dehydrogenase (fdhF) structural gene. Arch Microbiol. 1989;152(1):83–89. doi: 10.1007/BF00447016. [DOI] [PubMed] [Google Scholar]
- Scott D., Amy N. K. Molybdenum accumulation in chlD mutants of Escherichia coli. J Bacteriol. 1989 Mar;171(3):1284–1287. doi: 10.1128/jb.171.3.1284-1287.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperl G. T., DeMoss J. A. chlD gene function in molybdate activation of nitrate reductase. J Bacteriol. 1975 Jun;122(3):1230–1238. doi: 10.1128/jb.122.3.1230-1238.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart V., MacGregor C. H. Nitrate reductase in Escherichia coli K-12: involvement of chlC, chlE, and chlG loci. J Bacteriol. 1982 Aug;151(2):788–799. doi: 10.1128/jb.151.2.788-799.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart V. Nitrate respiration in relation to facultative metabolism in enterobacteria. Microbiol Rev. 1988 Jun;52(2):190–232. doi: 10.1128/mr.52.2.190-232.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tweedie J. W., Segel I. H. Specificity of transport processes for sulfur, selenium, and molybdenum anions by filamentous fungi. Biochim Biophys Acta. 1970 Jan 6;196(1):95–106. doi: 10.1016/0005-2736(70)90170-7. [DOI] [PubMed] [Google Scholar]
- Ugalde R. A., Imperial J., Shah V. K., Brill W. J. Biosynthesis of the iron-molybdenum cofactor and the molybdenum cofactor in Klebsiella pneumoniae: effect of sulfur source. J Bacteriol. 1985 Dec;164(3):1081–1087. doi: 10.1128/jb.164.3.1081-1087.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WILSON L. G., BANDURSKI R. S. Enzymatic reactions involving sulfate, sulfite, selenate, and molybdate. J Biol Chem. 1958 Oct;233(4):975–981. [PubMed] [Google Scholar]
- Yerkes J. H., Casson L. P., Honkanen A. K., Walker G. C. Anaerobiosis induces expression of ant, a new Escherichia coli locus with a role in anaerobic electron transport. J Bacteriol. 1984 Apr;158(1):180–186. doi: 10.1128/jb.158.1.180-186.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruijn F. J., Lupski J. R. The use of transposon Tn5 mutagenesis in the rapid generation of correlated physical and genetic maps of DNA segments cloned into multicopy plasmids--a review. Gene. 1984 Feb;27(2):131–149. doi: 10.1016/0378-1119(84)90135-5. [DOI] [PubMed] [Google Scholar]



