Abstract
We describe a method of designing artificial sequences that resemble naturally occurring sequences in terms of their compatibility with a template structure and its functional constraints. The design procedure is a Monte Carlo simulation of amino acid substitution process. The selective fixation of substitutions is dictated by a simple scoring function derived from the template structure and a multiple alignment of its homologs. Designed sequences represent an enlargement of sequence space around native sequences. We show that the use of designed sequences improves the performance of profile-based homology detection. The difference in position-specific conservation between designed sequences and native sequences is helpful for prediction of functionally important residues. Our sequence selection criteria in evolutionary simulations introduce amino acid substitution rate variation among sites in a natural way, providing a better model to test phylogenetic methods.
Computational protein design aims to identify sequences compatible with a desired structure or fold (1–4). Most design methods involve detailed energy functions with explicit modeling of protein structure at the atomic level and apply effective search algorithms (5, 6). They facilitate understanding of the physical and chemical principles governing protein structure and folding (4). Protein design can also be used to probe the sequence space (7), which has been applied in fold recognition (8). This idea can be extended to profile-based similarity searches, which derive a scoring function based on a multiple sequence alignment. Designed sequences resembling naturally occurring sequences could potentially be used to improve sequence profile, leading to more powerful homology detection. Sequence design can also be used in studying protein function and evolution (9, 10), as it is often related to evolutionary simulations. It is generally assumed that amino acid changes follow a stochastic process over long periods of time, and the fixation of substitutions is under evolutionary pressure to preserve protein activity. Evolutionary simulations can be made more realistic if structural and functional constraints are taken into account in the substitution process.
Knowledge-based approaches have been widely used to derive interaction potentials by statistical analysis of known protein structures (11, 12). Such potentials are used in various sequence design methods as stability constraints. Functional information about a protein family is embedded in naturally occurring homologs as positional amino acid conservation. Sequence profile, such as the position-specific scoring matrix generated by psi-blast (13), contains the positional conservation information. We attempt to introduce structural and functional constraints in sequence design by considering both pairwise interaction potentials and sequence conservation information.
Recently, a simulation-based design method (Z-score model) was used to study the protein evolutionary process (10). Structurally similar sequences are selected by minimizing the Z-score, which characterizes the energy gap between the native conformation and misfolded or unfolded conformations. Theory and folding simulations suggest that Z-score minimization can result in stable and fast-folding sequences under a random energy model (1, 14). This model was applied to study evolutionary time scales, substitution rates, and conservatism of protein fold families (10).
We use a Z-score design procedure to generate artificial sequences incorporating structural and functional information of naturally occurring sequences. Designed sequences represent an enlargement of sequence space around native sequences. We use designed sequences in profile-based sequence similarity searches. We also show that comparison of the conservation patterns of native sequences and designed sequences aids functional residue identification. By adding a Z-score criterion to evolutionary simulations, we introduce among-site rate variation in a natural way. We compare methods of evolutionary distance calculations under different evolutionary models.
Model and Methods
The Z-Score Model. In the Z-score model (1, 14), Monte Carlo simulations are performed to search for substitutions that favor the separation of the native-state energy (EN) from the average energy (〈E〉) of structurally unrelated conformations (decoys). The energy gap is characterized by the Z-score, defined as Z = (EN – 〈E〉)/σ(E), where σ(E) is the standard deviation of the energy of the decoys. The resulting change of Z-score (δZ) after an attempted substitution is calculated and the probability (P) to fix the substitution is guided by the Metropolis algorithm: P equals 1 if δZ is <0, otherwise P equals exp(–δZ/T). T is the parameter referred to as “temperature” that characterizes the tolerance to substitutions.
The Protein Energetic Model. Our scoring function is based on a simple energetic model that combines structural information and sequence conservation. The total energy (Et) of a protein structure is evaluated as a linear combination of a single-residue potential (Es) and a pairwise potential (Ep). The two potentials are related by a scaling factor w (weight): Et = wEp + (1 – w)Es, with the average value of 〈Et〉 = w 〈Ep〉 + (1 – w)〈Es〉 and standard deviation of  .
.
The single residue potential (Es) for the protein structure is derived from a multiple alignment of native sequences homologous to the target structure. Each position has a score contributing to the single-residue potential. The preference for amino acid ai in a position i is transformed to an empirical energy. For convenience, the psi-blast (13) score (Sai) of residue ai in position i is used and the single residue potential is Es =Σi Sai. The average value and standard deviation of the single residue potential are calculated as
 |
S0a is the average and σ(Sa) is the standard deviation of the psi-blast scores for residue type a for a random position. We estimate the average value and the standard deviation of psi-blast scores for each residue type from a statistical analysis of all positions with <50% gaps in 284 alignments in the smart database (15, 16).
The pairwise potential (Ep) is
 |
where M(ai, aj) is the Miyazawa-Jernigan contact energy (12) between residues ai and aj, Δij is the element of the contact matrix. The definition of a contact is in accord with what was used in deriving the Miyazawa-Jernigan potentials: if the centers of the two side chains are within 6.5 Å, Δij is 1, otherwise Δij is 0. We use a decoy model for the pairwise potential that has been described in ref. 10:
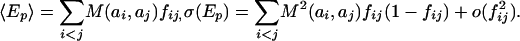 |
fij = f|i–j| = 〈Δij〉 is the frequency of a contact between positions i and j in structurally unrelated conformations and is estimated by a statistical analysis of known protein structures.
Weight Optimization. To find the optimal weight w between the single-residue potential and the pairwise potential, we compare the average Z-scores of native sequences to those of two randomized sets of sequences.
Native sequences have high Z-scores if only single-residue potential (sum of psi-blast scores) is used to evaluate the Z-scores (w = 0). Randomization of a native sequence eliminates the conservation patterns. Thus, the single-residue potential is effective in discriminating the native sequences from shuffled sequences (or shuffling along an alignment of native sequences horizontally). On the other hand, vertically shuffling each position in the alignment maintains the conservation patterns while eliminating the correlations between positions. The single-residue potential is not discriminative between such shuffled sequences and native sequences in terms of the average psi-blast scores. The pairwise potential should be effective if pairwise interactions between side chains can at least partially explain the covariation of amino acids between positions.
For a native alignment we perform two types of shuffling (“horizontally” along each sequence and “vertically” along each position) and compare the average Z-scores of the native sequences and the sequences in the shuffled alignment at varying values of w. We use the statistics of D = (Zn – Zs)/(σn + σs) to show the difference between native sequences and the horizontally or vertically shuffled sequences. Zn and Zs are average Z-scores of the native sequences and shuffled sequences, respectively; σn and σs are their standard deviations. D is scaled such that its value is between 0 and 1. Fig. 1 shows a typical diagram of the two types of difference statistics. We choose the cross point of the two curves as the optimal weight (w = 0.94), where both discriminations are close to the maximum value, i.e., 1. We performed weight optimization for various structures and found that the weight for the pairwise potential (w) was a value ≈0.9 in most cases. The scale of pairwise potentials (Miyazawa-Jernigan matrix elements) is ≈10-fold less than the scale of single residue potentials (psi-blast position-specific scoring matrix elements) such that at w = 0.9 the contributions of pairwise and single-residue potentials are about equal.
Fig. 1.
Weight optimization between pairwise potential and single-residue potential. The red line shows the average Z-score difference between native sequences and the random sequences by horizontal shuffling. The green line shows the average Z-score difference between native sequences and a random alignment resulted from vertical shuffling. The cross point (blue) depicts the optimal weight.
Designed Sequences Used in Homology Detection. We automate the sequence design procedure and similarity searches. For each template protein structure (taken from the Protein Data Bank), we perform psi-blast searches for homologous proteins for six iterations starting from the corresponding sequence (e-value cutoff 0.01). We obtain a position-specific scoring matrix (PSSM) using the –Q option in the program blastpgp (13). This PSSM is used for deriving the single residue potential. We obtain a multiple alignment of found homologs directly from the output of psi-blast. Seventy sequences are selected in the final alignment (if the sequence number is <70, then all of the sequences are included). We find the optimal weight between the two types of potentials by the sequence shuffling procedures described above or set the weight to be an empirical value of 0.9. We perform Monte Carlo simulations of the substitution process in the Z-score model starting from the initial protein sequence and structure. We collect designed sequences after a certain number of accepted substitutions of our simulations. We collect a set of designed sequences from a set of simulations started from different random numbers. We add the designed sequences to the native alignment and perform a new round of psi-blast searches starting from each individual sequence in the combined alignment and seeded with the combined alignment (-B option in the program blastpgp, e-value cutoff 0.01). We test different sets of designed sequences with different numbers of sequences (35, 70, and 140) and different substitution numbers (l/2, l, and 3l/2; l is the sequence length of the alignment). All found homologues are pooled together to form a set A.
As a control, we perform a psi-blast search to convergence starting from the initial sequence of the template structure. Found homologues are grouped by single-linkage clustering (the threshold of 1 bit per site) as implemented in the seals package (17), and the representative sequences are used as new queries for a new round of psi-blast searches. We select all found homologs to a set B. We compare sets A and B to check whether set A contains homologs not in B. We test the automated design and detection procedure on 48 oligonucleotide/oligosaccharide binding (OB)-fold domains, including cold shock protein 1mjc (the results are available in Table 2, which is published as supporting information on the PNAS web site, www.pnas.org).
We analyze the link between major cold shock protein and ribosomal protein S1 in greater detail. To obtain an alignment of high quality for 1mjc, we select representative sequences after clustering all find major cold shock homologs by using the blastclust program from the National Center for Biotechnology Information (sequence identity cutoff 70%). We align representative sequences by using t-coffee (18) followed by manual inspection and removal of short fragments, resulting in a curated alignment of 70 sequences. This alignment is used in design and homology detection.
Functional Residue Identification. For five protein families (trypsin, carboxypeptidase A, Rnase Sa, T4 lysozyme, and Rho termination factor), we select protein structures with and without ligands bound (see Table 1). For each structure with ligands, we define the active site zone (ASZ) to be all residues within 4.5 Å of the ligands. The ASZ is therefore enriched with residues important for function. We design 103 sequences with N = 10*l accepted steps for each structure without ligand(s) (l is the sequence length). We compute positional conservation in the native alignment and designed alignment from an entropy measurement with the Henikoff weighting scheme for amino acid frequency estimation (16, 19). We calculate and rank the conservation differences at each position between the native alignment and the designed alignment. For positions with the top 25% largest conservation decrease from the native to the designed alignment, we count the number belonging to the ASZ. We compare this number to the random expectation, assuming that functional positions do not exhibit significantly larger conservation decrease than nonfunctional positions (one-quarter of the total number of positions in the ASZ).
Table 1. Active site residues (ASR) with large conservation difference.
| Protein family | PDB with substrate | Apo PDB | Sequence length | No. of ASR | No. of ASR within the top 25% conservation difference | Expected no. |
|---|---|---|---|---|---|---|
| Carboxypeptidase A | 8cpa | 2ctn | 307 | 17 | 11 | 17*0.25 = 4.25 |
| Rho termination factor | 2a8v | 1a8v | 118 | 12 | 6 | 12*0.25 = 3 |
| Trypsin | 1btz | 2ptn | 223 | 21 | 12 | 21*0.25 = 5.25 |
| Rnase Sa | 1rge | 1rgg | 96 | 10 | 6 | 10*0.25 = 2.5 |
| T4 lysozyme | 1481 | 5lzm | 162 | 27 | 11 | 27*0.25 = 6.75 |
PDB, Protein Data Bank.
Evolutionary Distance Calculations. We study two evolutionary models. In the first model, we randomly select a position and attempted a substitution according to the Dayhoff PAM1 substitution probability matrix (20). Any attempted substitution is fixed. In the second model, we randomly choose a position and attempt a substitution similarly, but the fixation of the substitution is accordingtothe Z-score Metropolis criterion. We define one substitution cycle as the number of accepted substitutions equals the sequence length. The evolutionary distance (d) per substitution cycle is set to 1, that is, the average number of substitutions per site is 1. Starting from the initial sequence of 1mjc, we collect the final sequence after a certain number of substitution cycles (d). We calculate the normalized fraction of unchanged sites as q = (S – Srand)/(1 – Srand), where S is the identity of two sequences and Srand is the identity of two random sequences. We approximate the identity of two random sequences to be 0.05.
Results
Designed Sequences Improve Distant Homology Detection. Similarity search programs such as psi-blast (13) effectively use the information in alignments of native proteins. Designed sequences represent the expansion of the sequence space compatible with the fold and function of native sequences. To test whether designed sequences help homology detection, we add them to the native alignment and use the combined alignment to generate a profile for blast searches (psi-blast seeded with alignment).
We select OB-fold (21) structures for the test. OB-fold adopts a β-barrel structure consisting of five β strands. OB-fold is characteristic of a wide variety of proteins, most of which are involved in oligonucleotide or oligosaccharide binding. In the scop (Structural Classification of Proteins, version 1.57) (22) database, OB-fold is divided into seven superfamilies. The superfamily of nucleic acid-binding proteins is the most diverse and populated. Proteins within each superfamily are considered to be homologs. However, the OB-fold sequences can get so diverse that automatic psi-blast searches cannot link all of the proteins in the same superfamily. This provides us a good case for testing profile-based similarity searches. A total of 48 OB-fold domains are selected, and psi-blast searches are done with designed sequences added to the native alignments (see Model and Methods). For 14 domains, we find new hits compared with normal psi-blast searches with found homologs (see Table 2). Most hits are true positives as we manually confirm.
New hits are found in the superfamily of nucleotide-binding proteins. The most interesting case is the link between the major cold shock domain and ribosomal protein S1 domain. Both proteins belong to the cold-shock DNA-binding domain-like family according to scop. This family also includes other protein domains such as translational initiation factor 1 and Rho termination factor. However, automatic psi-blast searches (e-value cutoff 0.01, NR database in October 2001 with 764,279 sequences, 242,943,615 total letters) starting from the sequence of a major cold shock protein with known structure (Protein Data Bank ID code 1mjc) (23) converge within the major cold shock domains and could not make links to other domains in the same scop family. psi-blast searches seeded with a curated alignment (see Model and Methods) was carried out starting from each sequence in the alignment. Still no links between cold shock domain and other domains in the same family could be established (e-value cutoff 0.01). Using the structure of 1mjc (residues 4–70) and a curated alignment, we design 70 sequences with 67 accepted substitutions at the optimized weight. The 70 designed sequences are added to the native alignment, and a new round of psi-blast seeded with the combined alignment is performed. This time we detect two new homologues with e-value <0.01. One is ribosomal protein S1 from Rickettsia conorii [gene identification (gi) number: 15619842, best e-value 0.002], and the other is a hypothetical protein from Trypanosoma brucei (gi: 9366840, best e-value 0.004). Therefore, designed sequences aid identification of the remote homology relationship between the cold shock domain and S1 domain. Detection of R. conorii S1 protein is robust with regard to design parameters. It is consistently found with e-value >0.01 in most tested cases where the accepted substitution steps and/or the number of designed sequences added to the native alignment are varied (see Table 3, which is published as supporting information on the PNAS web site). Using a multiple alignment of major cold shock proteins directly from psi-blast output for design, we also detect the same S1 protein with e-value <0.01 (see Table 2).
We perform two control tests of alignment-seeded searches. In the first one, a random alignment generated by shuffling along each position of the native alignment is added to the native alignment. This procedure does not lead to detection of any new homologs, suggesting that the improvement of the profile by adding designed sequences is not caused by an increase of the alignment size with similar conservation properties. In the second control test the added sequences are obtained by simulating the evolutionary process under the Dayhoff PAM1 model (20). In this model, a site is chosen randomly, and the substitution is made according to the PAM1 substitution probability matrix. Unlike the Z-score design, there are no structural or functional constraints for substitution fixation. Alignments are generated with different parameters of design in accord with the above (number of accepted substitutions, number of designed sequences added to the native alignment). Still no new homologs are found. This result suggests that structural and evolutionary information used in the design procedure indeed plays an important role in improving profile-based similarity searches.
In Fig. 2, we plot a distance diagram illustrating the similarities among S1 sequences, major cold shock sequences, and the designed sequences (24). The S1 sequences (black circles) and major cold shock sequences (red circles) form two distinct clusters with no overlapping between them, suggesting that the similarities between the two groups are low. The designed sequences (blue circles) all cluster around the native major cold shock sequence of 1mjc (the yellow point), because they are generated by limited number of substitutions made from it (on average one substitution per site). Some designed sequences are closer to the R. conorii S1 protein (green circle) than most of the native sequences. This may be the reason for an improved sequence profile that leads to the detection of this S1 protein by adding designed sequences.
Fig. 2.
Distance diagram showing the similarities among major cold shock sequences, designed sequences, and ribosomal S1 sequences.
Design and Functional Residue Identification. It is well known that many proteins trade stability for function (25). Residues important for catalysis or molecular interactions are often not optimized for stability. Their conservation mainly reflects the functional constraints. There are also positions where conservation is caused mainly by stability constraints. Although conservation is widely used to indicate functionality (16), it is not obvious that it discriminates positions with mainly functional constraints from positions with mainly structural constraints. In most cases, the former is of the most interest to the study of protein activities.
In our design scheme, single-residue potential (profile scores) characterizes the conservation properties of naturally occurring sequences. If only single-residue potential is used (w = 0) in the design process, the conservation pattern of the native alignment will be largely maintained in the designed sequences. Pairwise potential reflects physical interactions between residues in contact and only exerts stability constraints in the design. We expect that incorporation of pairwise potential in design tends to maintain the conservation of positions contributing to structural stability while weakening the conservation of functional residues in the designed alignment. We select five well-studied protein families to test this idea. For a representative structure of each family, we design 103 artificial sequences with the optimized weight. The differences of conservation at each position between the native alignments and the designed alignments are measured and ranked in a descending order. Table 1 shows that residues belonging to the ASZ tend to have larger conservation decreases than other part of the protein.
The conservation difference measure could be useful for predicting functional residues on a protein structure. Fig. 3 illustrates the active site of trypsin. If conservation of the native alignment is mapped onto the protein structure, all conserved positions important for stability or function are highlighted (red), for instance, the two disulfide bond-forming cysteines and the catalytic triad (with side chains shown). If the conservation difference between the native alignment and the designed alignment is mapped onto the structure, the functional catalytic triad are still highlighted because they have large conservation change. The conservation of the two cysteines is maintained in the designed alignment. They are not highlighted in Fig. 3 because changes in their conservation are small. This example demonstrates that conservation difference could be a better measure in pinpointing positions with mainly functional constraints than simple conservation values.
Fig. 3.
(a) Ribbon diagrams showing sequence conservation in a multiple alignment of native trypsin sequences. (b) Ribbon diagrams showing sequence conservation difference between native alignment and the designed alignment of trypsin family. Red and blue correspond to the highest and the lowest conservation or conservation differences, respectively.
Testing Evolutionary Distance Estimators. Simulations of evolutionary process are widely used to test phylogenetic methods such as evolutionary distance estimation and tree reconstruction (26, 27). Our design procedure can be viewed as a simulation of evolutionary process in which stability and function are taken into account in the selective fixation of substitutions. Because different sites are under different selection pressure, rate variations among sites are naturally introduced in our model.
Here we use our design procedure to test different methods of estimating evolutionary distances (d) from sequence similarity. Sequence similarity is quantified by the normalized fraction of unchanged positions: q = (S – Srand)/(1 – Srand). Two models of substitution process are tested. One simulation process is under a PAM1 model with equal probability of substitution attempt at each position and no selection pressure of substitution fixation. Under this model we expect that the substitution rates do not vary among sites. In the other model the Z-score selection criterion is added to the PAM1 model (see Model and Methods), and the substitution rates could vary among sites because of structural and functional constraints. The relationships of sequence similarity and evolutionary distance are shown in Fig. 4. It is clear that with structural and functional constraints (red line), the normalized fraction of unchanged positions decreases more slowly with the increase of evolutionary distance than that under the equal-rate PAM1 model without selection on substitution fixation (black line). Two formulae for estimating the relationship between q and d are also plotted. One is the Poisson relationship: q = exp(–d) (blue line), derived under the assumptions that all residues in a sequence have the same probability of change and every amino acid has the same probability of changing to other amino acids. It fits very well to the PAM1 model without structural and functional selection pressure. The other formula is the one of the formulae Grishin proposed: q = 1/(1 + d) (green line) (28), which is derived assuming that the substitution rates among sites follow an exponential distribution. The result of simulation with Z-score model is closer to what this formula predicts.
Fig. 4.
Plot of the relationship of evolutionary distance vs. normalized sequence identity. The red curve is for simulation under the Dayhoff model with a Z-score criterion. The black curve is for simulation under the Dayhoff model without a Z-score criterion. The green curve is for the Grishin's formula: q = 1/(1 + d). The blue curve is for the formula under the Poisson assumption: q = exp(–d).
Discussion
Scoring Function. Most protein design efforts involve detailed energy functions to model the forces of interactions at the atomic level. Even under the approximations of fixed backbone and discrete rotamer conformations (29), the search space is quite large (4). Because of such limitations as the inaccuracy in the energy functions and the time complexity, it is not feasible to model the detailed structural differences among naturally occurring homologs or the detailed structural changes during a trajectory of natural substitutions. Instead, we select a coarse-grained energy model. Under such a model, foldablility and stability cannot be guaranteed. Rather, the designed sequences should be deemed as artificial sequences with native-like conservation properties. In homology detection tests we have limited the substitution steps to be around one substitution per site, so that not too many deleterious substitutions are introduced.
We use a scoring function that is a linear combination of the pairwise interaction potential between positions and the observed conservation for each position. Similar combination of energy terms has been used in fold recognition (30, 31), ab initio folding (32) and protein design efforts (33). Both energy terms of the scoring function are simple to calculate. The Miyazawa-Jernigan energy terms (12) are derived from a statistical analysis of side-chain contacts in known structures. Our pairwise potential only depends on the side-chain contact patterns and omits the interaction details such as the packing of the side chains. We assume that the contact patterns are largely maintained in structurally similar proteins or during the substitution process. The single-residue potential is derived from a multiple alignment of native sequences and manifests the fitness of amino aids at a position. It contains various structural information, such as backbone-dependent amino acid preferences, exposure to the solvent and, indirectly, pairwise side-chain interactions. The single residue potential also introduces functional and evolutionary constraints to make the designed sequences more native like. These constraints should be helpful in homology detection. There is certainly overlapping between the two terms in a linear combination, which is a common problem in scoring functions used in protein design. We propose a method of finding the optimal scaling factor between the two terms by comparing native sequences with two sets of shuffled sequences.
Artificial Sequences for Similarity Searches. The most sensitive similarity search tools effectively use evolutionary information in the form of a position-specific scoring scheme. Profiles (34), transformed into position-specific scoring matrices (13) or encoded in hidden Markov models (35), are derived from alignments of homologous proteins. The quality of a profile depends on the diversity of the sequences and the quality of the multiple alignment. psi-blast (13) uses an iterative procedure to include newly found homologs to improve the quality of the profile. Alignments of better quality can help homology detection (36). We try to improve the quality of the profile by integrating structural information from available 3D structures and functional information from multiple alignments. The artificial sequences produced by the design procedure represent enlargement of the sequence space around naturally occurring sequences. Inclusion of designed sequences to the native alignment led to improvement of the statistical significance of the similarity search results between major cold shock domain and ribosomal S1 domain. New homologues were also found for other OB-fold domains by adding designed sequences to the native alignment. For each OB-fold structure, we tried adding 35, 70, and 140 designed sequences to the native alignment. Adding more designed sequences did not help finding more new homologs, probably because more deleterious substitutions were introduced to the alignment.
However, we also notice several limitations in this homology detection approach by adding artificial sequences. First, the scoring function is too coarse-grained in characterizing the compatibility of the sequence with the fold. For this reason, we start substitution process from the native sequence with the structure and do not let the substitution process go too far. This leads to a limited change of the profile after adding the designed sequences to the native alignment. Too many substitutions can make the profile worse and could possible introduce false positives in similarity searches, as we find in the tests of OB-fold proteins. Second, other evolutionary events such as insertions and deletions are not modeled in the simulation. Third, the single residue potential is derived from the native alignment. If only several very close homologs are identified by psi-blast searches, the information content of the native alignment will be low (as is the case for many OB-fold domains not from the superfamily of nucleotide-binding proteins). Fourth, the existence and the distribution of distant homologs in sequence space are unpredictable, so are the structural and functional changes in them. Designed sequences may not vary in the direction needed to find these homologs. For 48 OB-fold structures we tried, new homologues were detected for only 14 of them. Despite these obvious limitations, the link between cold shock protein and S1 protein suggests that our design procedure can be useful in making discoveries of remote homologs.
Functional Constraints and Stability Constraints in Design. The two most important constraints for sequence conservation are stability and functionality. Conservation caused by stability constraints and functional constraints can overlap, but usually only one type of constraint dominates in a conserved position. Functional residues, involved in catalysis or interactions with other molecules, are often positioned on the surface of a protein. In fact, they are often not optimized for stability (25). On the other hand, stability is the dominant constraint for the residues in hydrophobic cores. In our design procedure, the single residue potential captures mixed evolutionary information about conservation whereas the pairwise potential only captures stability information. Incorporation of pairwise potential tends to weaken the conservation of functional positions not under much stability constraints. Thus, functional positions tend to show large conservation differences between native and designed alignments. Because in most cases we are more interested in functional positions than positions under stringent stability constraints, analyzing the conservation difference of the native and designed alignment could be useful for prediction of functional residues.
Design as a Simulation of Evolutionary Process. The simulation of the protein evolutionary process is widely used in various phylogenetic tests. Simulations can be done under different models of substitution events. The simplest model is the Poisson model that assumes equal probability of change at each position in a sequence and equal probability of change among different amino acid types. Other models may take into account the likelihood variation of substitutions among amino acids by using a substitution probability matrix (20) or the variation of substitution rates among sites (27). Substitution matrices such as the PAM series matrices derived by Dayhoff et al. (20) usually come from a statistical analysis of substitution events in homologous proteins and manifest the general properties of exchange among amino acids (e.g., hydrophobic or hydrophilic preferences). Variation of substitution rates among sites is usually approximated by a certain type of statistical distribution such as the γ distribution (37). However, for a particular protein family, the substitution events not only follow the general substitution tendencies among amino acids but also are influenced by the structural and functional properties specific for this family. Selective fixation of substitutions is family specific to maintain the protein's structure and function.
Our design procedure can be used as a simulation of the evolutionary process. The fixation of substitutions is under selection by the functional and structural constraints of a protein family, rendering this type of simulation more realistic. The variation of substitution rates among sites is caused by these constraint. We show that the formula q = 1/(1 + d) (28) that takes into account the rate variability among sites approximates better the empirical relationship observed in our evolutionary simulation. This type of simulation should also be useful in testing other methods in phylogeny, such as tree-building methods.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant GM52126 (to E.I.S.) and National Institutes of Health National Research Service Award Fellowship GM20251 (to N.V.D.).
Abbreviations: OB, oligonucleotide/oligosaccharide binding; ASZ, active site zone.
References
- 1.Shakhnovich, E. I. & Gutin, A. M. (1993) Protein Eng. 6, 793–800. [DOI] [PubMed] [Google Scholar]
- 2.Shakhnovich, E. I. (1998) Folding Des. 3, R45–R58. [PubMed] [Google Scholar]
- 3.DeGrado, W. F., Summa, C. M., Pavone, V., Nastri, F. & Lombardi, A. (1999) Annu. Rev. Biochem. 68, 779–819. [DOI] [PubMed] [Google Scholar]
- 4.Pokala, N. & Handel, T. M. (2001) J. Struct. Biol. 134, 269–281. [DOI] [PubMed] [Google Scholar]
- 5.Dahiyat, B. I. & Mayo, S. L. (1997) Science 278, 82–87. [DOI] [PubMed] [Google Scholar]
- 6.Kuhlman, B. & Baker, D. (2000) Proc. Natl. Acad. Sci. USA 97, 10383–10388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koehl, P. & Levitt, M. (2002) Proc. Natl. Acad. Sci. USA 99, 1280–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koehl, P. & Levitt, M. (2002) Proc. Natl. Acad. Sci. USA 99, 691–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raha, K., Wollacott, A. M., Italia, M. J. & Desjarlais, J. R. (2000) Protein Sci. 9, 1106–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dokholyan, N. V. & Shakhnovich, E. I. (2001) J. Mol. Biol. 312, 289–307. [DOI] [PubMed] [Google Scholar]
- 11.Sippl, M. J. (1990) J. Mol. Biol. 213, 859–883. [DOI] [PubMed] [Google Scholar]
- 12.Miyazawa, S. & Jernigan, R. L. (1999) Proteins 34, 49–68. [DOI] [PubMed] [Google Scholar]
- 13.Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997) Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shakhnovich, E. I. & Gutin, A. M. (1993) Proc. Natl. Acad. Sci. USA 90, 7195–7199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schultz, J., Copley, R. R., Doerks, T., Ponting, C. P. & Bork, P. (2000) Nucleic Acids Res. 28, 231–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pei, J. & Grishin, N. V. (2001) Bioinformatics 17, 700–712. [DOI] [PubMed] [Google Scholar]
- 17.Walker, D. R. & Koonin, E. V. (1997) Intelligent Syst. Mol. Biol. 5, 333–339. [PubMed] [Google Scholar]
- 18.Notredame, C., Higgins, D. G. & Heringa, J. (2000) J. Mol. Biol. 302, 205–217. [DOI] [PubMed] [Google Scholar]
- 19.Henikoff, S. & Henikoff, J. G. (1994) J. Mol. Biol. 243, 574–578. [DOI] [PubMed] [Google Scholar]
- 20.Dayhoff, M. O., Schwartz, R. M. & Orcutt, B. C. (1978) in Atlas of Protein Sequences and Structures, ed. Dayhoff, M. O. (National Biomedical Research Foundation, Washington, DC), Vol. 5, Suppl. 3, pp. 345–352. [Google Scholar]
- 21.Murzin, A. G. (1993) EMBO J. 12, 861–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murzin, A. G., Brenner, S. E., Hubbard, T. & Chothia, C. (1995) J. Mol. Biol. 247, 536–540. [DOI] [PubMed] [Google Scholar]
- 23.Schindelin, H., Jiang, W., Inouye, M. & Heinemann, U. (1994) Proc. Natl. Acad. Sci. USA 91, 5119–5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grishin, V. N. & Grishin, N. V. (2002) Bioinformatics 18, 1523–1534. [DOI] [PubMed] [Google Scholar]
- 25.Shoichet, B. K., Baase, W. A., Kuroki, R. & Matthews, B. W. (1995) Proc. Natl. Acad. Sci. USA 92, 452–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adell, J. C. & Dopazo, J. (1994) J. Mol. Evol. 38, 305–309. [DOI] [PubMed] [Google Scholar]
- 27.Feng, D. F. & Doolittle, R. F. (1997) J. Mol. Evol. 44, 361–370. [DOI] [PubMed] [Google Scholar]
- 28.Grishin, N. V. (1995) J. Mol. Evol. 41, 675–679. [DOI] [PubMed] [Google Scholar]
- 29.Dunbrack, R. L., Jr., & Karplus, M. (1993) J. Mol. Biol. 230, 543–574. [DOI] [PubMed] [Google Scholar]
- 30.Panchenko, A. R., Marchler-Bauer, A. & Bryant, S. H. (2000) J. Mol. Biol. 296, 1319–1331. [DOI] [PubMed] [Google Scholar]
- 31.Xu, Y. & Xu, D. (2000) Proteins 40, 343–354. [PubMed] [Google Scholar]
- 32.Simons, K. T., Kooperberg, C., Huang, E. & Baker, D. (1997) J. Mol. Biol. 268, 209–225. [DOI] [PubMed] [Google Scholar]
- 33.Voigt, C. A., Gordon, D. B. & Mayo, S. L. (2000) J. Mol. Biol. 299, 789–803. [DOI] [PubMed] [Google Scholar]
- 34.Gribskov, M., McLachlan, A. D. & Eisenberg, D. (1987) Proc. Natl. Acad. Sci. USA 84, 4355–4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eddy, S. R. (1998) Bioinformatics 14, 755–763. [DOI] [PubMed] [Google Scholar]
- 36.Abbott, J. J., Pei, J., Ford, J. L., Qi, Y., Grishin, V. N., Pitcher, L. A., Phillips, M. A. & Grishin, N. V. (2001) J. Biol. Chem. 276, 42099–42107. [DOI] [PubMed] [Google Scholar]
- 37.Guindon, S. & Gascuel, O. (2002) Mol. Biol. Evol. 19, 534–543. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.






