Abstract
Single-nucleotide polymorphisms (SNPs) constitute the bulk of human genetic variation and provide excellent markers to identify genetic factors contributing to complex disease susceptibility. A rapid, sensitive, and inexpensive assay is important for large-scale SNP scoring. Here we report the development of a multiplex SNP detection system using silicon chips coated to create a thin-film optical biosensor. Allele-discriminating, aldehyde-labeled oligonucleotides are arrayed and covalently attached to a hydrazinederivatized chip surface. Target sequences (e.g., PCR amplicons) then are hybridized in the presence of a mixture of biotinylated detector probes, one for each SNP, and a thermostable DNA ligase. After a stringent wash (0.01 M NaOH), ligation of biotinylated detector probes to perfectly matched capture oligomers is visualized as a color change on the chip surface (gold to blue/purple) after brief incubations with an anti-biotin IgG-horseradish peroxidase conjugate and a precipitable horseradish peroxidase substrate. Testing of PCR fragments is completed in 30–40 min. Up to several hundred SNPs can be assayed on a 36-mm2 chip, and SNP scoring can be done by eye or with a simple digital-camera system. This assay is extremely robust, exhibits high sensitivity and specificity, and is format-flexible and economical. In studies of mutations associated with risk for venous thrombosis and genotyping/haplotyping of African-American samples, we document high-fidelity analysis with 0 misassignments in 500 assays performed in duplicate.
The human genome contains nucleotide sequence variations in different individuals at an average frequency of 0.1% (1). Single nucleotide changes, variation in copy number of di-, tri-, or tetranucleotide repeats, and small deletions or insertions all contribute to overall genomic diversity. Because of the sheer abundance of single-nucleotide polymorphisms (SNPs), considerable effort has been expended to identify >2 million SNP loci that span the entire human genome (ref. 2 and National Center Biotechnology Information Single Nucleotide Polymorphism Database, www.ncbi.nlm.nih.gov/SNP). SNPs have significant diagnostic potential, and SNP analysis provides a theoretical basis for performing candidate gene or whole-genome association studies for common complex (polygenic) diseases (3, 4).
A large variety of different techniques has been developed for SNP scoring, each of which has specific advantages and/or limitations (reviewed in refs. 1, 5, and 6). Different strategies for SNP allele discrimination include restriction-enzyme digestion (7), allele-specific hybridization (8), primer extension by nucleotide incorporation (9), invasive nuclease cleavage (10), oligonucleotide ligation (11), mass spectroscopy (12), and DNA sequencing of PCR amplification products (13). Although a wide range of detection modalities are used, the most commonly used are fluorescence-based, including the Third Wave Invader (14) and Applied Biosystems TaqMan (15) assay systems, considered by many to be the industry “gold standards.” Unfortunately, most SNP assay methods suitable for high-throughput analysis require expensive instrumentation for their implementation. A genotyping procedure that is inexpensive, flexible enough to accommodate low-, moderate-, or high-throughput needs, takes minimal effort for probe design, and provides rapid SNP scoring with high sensitivity and specificity with little if any instrumentation requirements would be desirable. Here we describe a DNA ligation-based SNP assay conducted on an optical thin-film biosensor chip that meets these criteria. This method will have multiple uses in such areas as determining the correlation between genotype and disease susceptibility (and hence perhaps providing hints to pathogenic mechanisms), genetic epidemiology and population genetics, and diagnosis of infectious diseases (including discrimination between two closely related disease agents with differing virulence).
Methods and Materials
The Thin-Film Biosensor Chip. Thin-film biosensors are capable of transducing specific molecular interactions into signals that can be visualized by the naked eye because mass deposited on the thin-film surface by enzymatic catalysis alters the wavelength of light reflected by the optical layer resulting in a perceived color change on the surface (16). The biosensors used here were prepared by coating the surface of 10-cm-diameter silicon wafers with a 475-Å layer of silicon nitride (Si3N4), which serves as the optical layer. To facilitate covalent attachment of biomolecules, the wafers also were spin-coated with a 135-Å layer of T structure aminoalkylpolydimethylsiloxane (TSPS) and poly(Phe-Lys) passively adsorbed to the TSPS layer (17, 18). The wafers were cut into 6 × 6-mm squares by using a laser knife, with each square constituting an assay chip. These chips, commercially available from ThermoBiostar (Louisville, CO), were generously provided by the company and used for individual assays or assembled as arrays in a 96-square-well microtiter plate. The ε-amino amino groups of the poly(Phe-Lys) were converted to hydrazines as described below to optimize the simultaneous printing of multiple oligonucleotide arrays.
Modification of Chip Surface and Oligonucleotide-Coupling Chemistry. Poly(Phe-Lys)-coated wafers were incubated in a 25-ml solution of 1–10 μM succinimidyl hydrazinium nicotinate hydrochloride (Solulink, San Diego) in 0.1 M sodium borate buffer, pH 8.1–8.4 for 2 h followed by washing in distilled H2O. Oligonucleotides were chemically synthesized on a standard instrument (Applied Biosystems) incorporating a 5′-aldehyde-substituted phosphoramidite (Solulink) into the synthetic process. Both the aldehyde- and hydrazine-functionalized molecules are chemically stable for many months in aqueous solutions when stored separately, but they react rapidly when mixed to form stable hydrazones. Aldehyde-labeled oligonucleotides were diluted to different concentrations in 0.1 M sodium phosphate buffer, pH 7.8, and 1–200 nl was spotted, depending on the printing method, onto hydrazine-coated chips. After incubation at room temperature for 2 h, chips were rinsed with distilled H2O, incubated with 0.1% SDS at 60°C for 2 h, then washed with H2O, and air-dried. Spotted chips can be stored at room temperature for at least 6 months before use.
Aldehyde-labeled chips [prepared by soaking wafers for 2 h in a solution of 1–10 μM succinimidyl-4-formyl benzoate (Solulink)/0.1 M sodium borate buffer, pH 8.2] work equally well when spotted with 5′-hydrazine-labeled oligomers. The latter are prepared by reacting 5′-amino-labeled oligomers with a 10-fold molar excess of succinimidyl hydrazinium nicotinate hydrochloride for 2 h in 0.1 M sodium borate buffer, pH 8.2, followed by purification by using a gel-filtration G-25 spin column (Sephadex, Sigma–Aldrich).
Oligonucleotide Synthesis. For each selected SNP site, a pair of allele-discriminating oligonucleotide probes (P-1 oligos) and a single detector oligonucleotide (P-2) is designed. The P-1 oligos have 5′-aldehyde groups, 10 deoxyadenosine residues at their 5′ ends that constitute a “spacer,” followed by 40 nucleotides complementary to the corresponding SNP target sequence. These P-1 capture probes differ only at their 3′-terminal nucleotide, each matching one of the two SNP genotypes. The P-2 detector probe, containing ≈20 nucleotides with sequence immediately adjacent to the SNP nucleotide, is synthesized with a biotin at its 3′ end for detection and phosphate at its 5′ end for ligation. SNP oligomer sequences, PCR primer sequences, and PCR amplification conditions are listed in Tables 1–3 (which are published as supporting information on the PNAS web site, www.pnas.org), respectively. Oligonucleotides can be used without postsynthesis purification, thus reducing probe-production costs.
Preparation of PCR Amplicons. Genomic DNA (50 ng) of each individual was PCR-amplified with single or multiplex (three to five times) PCR primer sets in 20-μl reactions with 1.0 unit of AmpliTaq Gold (Hoffmann–LaRoche). The concentrations of PCR products, measured by a PicoGreen double-stranded DNA quantitation kit (Molecular Probes), generally were 20–50 ng/μl (≈1010 molecules per μl). PCR fragment size, between 200 and 500 bp, was confirmed by agarose gel electrophoresis. In the study of the African-American samples, PCR products from 20 chromosome 17 markers from one individual were pooled, and 20 μl of pooled product was added to 100 μl of standard hybridization/ligation solution (see Standard SNP Assay Protocol). The solution was heated at 95°C for 3 min, cooled to 60°C, and added directly to a microtiter plate well containing a biosensor chip. The chips were processed and digital images acquired as described below.
Standard SNP Assay Protocol. P-1 capture probes are spotted by manual pipette, a robotic Hamilton pipetting device, or a microarray printer by using 200, 40, or 1 nl per spot, respectively, onto biosensor chips from 1.0 μM oligonucleotide stocks in 0.1 M sodium phosphate buffer, pH 7.8. Multiple PCR amplicon targets, from 100 to 500 bp in length, at a concentration of 100 fmol each per 100-μl reaction are denatured and hybridized on the chip in the presence of 10 nM biotin P-2 probe (one for each SNP site) and 5 units of a Thermus thermophilus DNA ligase (Ampligase, Epicentre Technologies, Madison, WI) in 20 mM Tris·HCl, pH 8.3/25 mM KCl/10 mM MgCl2/0.5 mM nicotinamide adenine dinucleotide/0.01% Triton X-100/5 mg/ml acid-treated casein. Recent experiments indicate that the Lys-294 → Arg mutant of T. thermophilus ligase (19) gives 10-fold better allele discrimination than the wild-type enzyme, and it is now included as a standard assay reagent (this enzyme is not commercially available yet). The hybridization/ligation reaction is incubated for 20 min at 60°C. Single or multiple chips, i.e., in a 96-well plate, can be processed simultaneously. After a stringent wash (0.01 M NaOH, for 10–20 sec at room temperature) and a rinse in 0.1× standard saline citrate (1× SSC = 0.15 M sodium chloride/0.015 M sodium citrate, pH 7), the chips are incubated with an anti-biotin IgG-horseradish peroxidase (HRP) conjugate (Jackson ImmunoResearch; 1:1,000 dilution from a 1 mg/ml stock in a buffer containing 5× SSC and 5 mg/ml acid-treated caseine) for 10 min. The chips are rinsed with 0.1× SSC and 100 μl of a tetramethyl benzidine formulation from BioFx Laboratories (Owings Mills, MD) is added to the chips and incubated for 5 min at room temperature, then rinsed in ddH2O, air-dried, and analyzed. At lower spotting density SNPs can be scored readily by eye with or without a magnifier; however, chips spotted at high oligonucleotide density >32/36 mm2 are best visualized via digital images. For this we used a simple dissection microscope (Leica, Wild Model M10) fitted with an inexpensive digital camera (Sony Photo camera model DKC-ST5) with chip illumination by a standard tensor lamp. Densitometric analysis of digital images provides quantitative data on signal intensity with a dynamic range of ≈2 logs.
Results
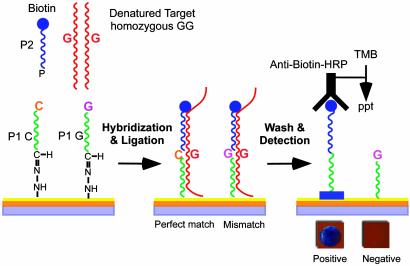
Strategy for SNP Genotyping on a Thin-Film Biosensor Chip. The SNP assay described here utilizes ligation of allele-discriminating oligonucleotides covalently arrayed on thin-film biosensor silicon chips (see Methods and Materials and Fig. 1). For each SNP to be interrogated, one pair of allele-discriminating oligomers (P-1 capture probes), differing only in their 3′-terminal nucleotide sequence, are covalently attached by their 5′ termini to the chip surface. A second oligonucleotide probe (P-2) with sequence immediately adjacent to the SNP nucleotide carries a biotin at the 3′ end for detection and a phosphate at its 5′ end for ligation. Target DNA hybridization and P-1–P-2 ligation reactions are done simultaneously during a 20-min incubation in the presence of a thermostable DNA ligase. After a stringent wash with NaOH to remove all nonligated molecules, immobilized biotinylated P-2 is detected by incubation with an anti-biotin IgG-HRP conjugate and a precipitable HRP substrate. Mass deposition from the substrate makes a distinguishable color change from gold to blue/purple on the chip surface, which, depending on the oligonucleotide spotting density, can be read either by eye or a simple digital-imaging system.
Fig. 1.
Strategy for SNP detection on thin-film biosensor chips (see text). TMB, Tetramethylbenzidine.
Modification of Chip Surface and Oligonucleotide Chemistry for Better Immobilization of Capture Probes. Reproducible immobilization of capture oligonucleotide probes is critical to chip-production consistency. In our initial experiments, 5′ amino-terminal modified oligonucleotides were reacted with a bis-N-hydroxysuccinamide ester cross-linking agent, and the mixture was spotted onto poly(Phe-Lys)-coated chips as described (18). Although this proved satisfactory for producing a small number of chips with only a few oligonucleotides arrayed on each, the amount of chip-bound oligomer decreased significantly when printing runs were longer than a few hours, presumably due to the hydrolysis of the bis-N-hydroxysuccinamide ester in aqueous solution. Thus the quality of chips was highly variable. We therefore evaluated several other types of coupling chemistries. The most robust and reproducible method used hydrazine–aldehyde interactions that form stable hydrazone linkages between the chip surface and capture oligonucleotides.
The preparation of hydrazine-derivatized chips, 5′-aldehydelabeled P-1 oligos, and conditions for the covalent linkage of oligomers to the chip surface are described in Methods and Materials. Molecules containing aldehyde (R—CHO) or hydrazine groups (R-NH·NH2) are stable in aqueous solutions for months when stored separately; however, after mixing they react rapidly to form stable hydrazones (R1—NH—N CH—R2). This reaction chemistry is quite specific: Hydrazines do not react with primary amines, carboxyl, or sulfhydryl groups; although aldehydes will react with primary amines, the resultant Schiff's bases are unstable and require reduction for stabilization. The use of this coupling chemistry routinely gave highly reproducible oligonucleotide arrays with uniform concentrations of oligomer on each spot even with spotting runs ≥24 h.
CH—R2). This reaction chemistry is quite specific: Hydrazines do not react with primary amines, carboxyl, or sulfhydryl groups; although aldehydes will react with primary amines, the resultant Schiff's bases are unstable and require reduction for stabilization. The use of this coupling chemistry routinely gave highly reproducible oligonucleotide arrays with uniform concentrations of oligomer on each spot even with spotting runs ≥24 h.
Defining Sensitivity and Specificity Parameters of Biosensor Chips. Various lengths of P-1 (20-, 30-, 40-, and 50-mer) and P-2 (10-, 12-, 14-, 16-, 18-, 20-, 22-, and 24-mer) probes were initially tested for their ability to discriminate alleles robustly, and it was determined that 40-mer P-1 probes and 20-mer P-2 probes provided optimal specificity and sensitivity on the biosensor chip (data not shown). We next established the concentration of P-1 capture probes required to detect defined amounts of PCR target sequences with short (≤20-min) hybridization/ligation times. Because P-1 probes can be spotted on a 36-mm2 chip at low density by hand or at moderate to high density by robotic printing devices, it was important to determine how spotting density affected detection sensitivity.
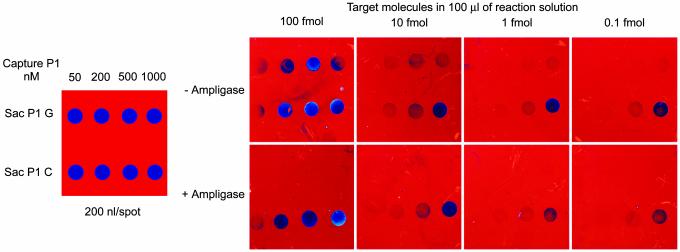
Allele-discriminating P-1 probes for the “Sac” (G/C) SNP (rs4646993) at the human HOXB6 gene were spotted manually at 10, 40, 100, and 200 fmol of P-1 per spot in a total volume of 200 nl. The chips were hybridized for 20 min at 60°C with different concentrations (0.1–100 fmol) of PCR product (560 bp) from a homozygous (G, G) individual in the presence of biotinylated Sac SNP P-2 probe (10 nM). One set of chips was subjected to hybridization without DNA ligase; these were washed posthybridization with 0.1× SSC at room temperature. A second set of chips were hybridized in the presence of T. thermophilus DNA ligase (Ampligase, 5 units per reaction) such that ligation reactions could be done simultaneously; these chips were washed with 0.01 M NaOH and then with 0.1× SSC, both at room temperature. Binding and/or ligation of biotinylated P-2 probe was visualized by a 10-min incubation with an anti-biotin IgG-horseradish (HRP) conjugate followed by a 5-min incubation with tetramethylbenzidine, a precipitable HRP substrate that increases thin-film thickness, thereby inducing the perceived color change. The chips then were washed, dried, and digitally imaged. Representative images are shown in Fig. 2.
Fig. 2.
Determination of allele discrimination specificity and target DNA-detection sensitivity on the biosensor chip. Chips were spotted manually with 200 nl of the HoxB6 gene Sac (G/C) SNP P-1 probes at the concentrations indicated and hybridized to target DNA (a 560-bp PCR product) at concentrations of 100, 10, 1, and 0.1 fmol in the presence of the biotinylated SAC P-2 probe with and without DNA ligase. The–Ampligase chips were assayed posthybridization with no 0.01 M NaOH wash step, whereas the +Ampligase chips were NaOH-treated before visualization.
First, as expected, in the absence of ligation no appreciable allele discrimination was observed (–Ampligase chips), particularly at the higher PCR target concentrations. Second, excellent allele discrimination is obtained when hybridization is combined with Ampligase ligation at all target sequence concentrations used; signal is detected only on the P-1 C probe that perfectly matches the homozygous (G, G) target. Third, although the signal intensity decreases as the concentration of target sequence is lowered, as little as 0.1 fmol of target (6 × 107 molecules) can be detected if the P-1 probe is spotted at 200 fmol per spot. Fourth, comparison of signal intensity of chips with and without ligation demonstrates that the ligation efficiency is quite high (≥50%).
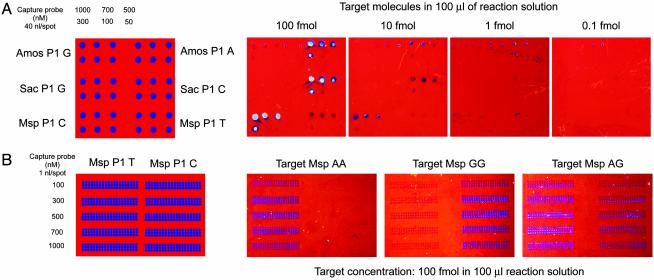
The effects of spot size (and therefore the number of spots per chip) and P-1 probe concentration on the sensitivity of target DNA detection was examined next (Fig. 3). Spotting of P-1 probes from stock solutions >1 μM did not increase detection sensitivity further (data not shown). However, as expected, the lower limit of detection decreased when smaller amounts of capture oligomer were spotted. Results of hybridization of target DNAs to P-1 capture probes ranging in concentration from 2 to 40 fmol per 40-nl spot (spotted with a pipetting robot) are shown in Fig. 3A. The P-1 probes are for three different SNP sites within the HOXB gene cluster (“Amos” G/A, Top; Sac G/C, Middle; “Msp” C/T, Bottom). Target DNA (a mixture of PCR products from TT, GG, and GG homozygous individuals at the Amos, Sac, and Msp sites, respectively) was hybridized at concentrations of 100, 10, 1.0, and 0.1 fmol per 100-μl reaction in the presence of Ampligase. Although no signal was observed by using 0.1 fmol of target DNA at any P-1 capture probe concentration, very strong signals (silver-to-purple) were seen by using 40–100 fmol of target DNA on spots containing 20–40 fmol of complementary P-1 capture probe. Considerably lighter signals were visible on the Amos P-1-G and Msp P-1-T probes, indicating some ligation to the mismatch probe.
Fig. 3.
Specificity and sensitivity of SNP genotyping on biosensor chips spotted with 40 nl (A) or 1 nl (B) of P-1 capture probe at the indicated concentrations. (A) Oligomers for three different HoxB gene SNPs were arrayed and hybridized with the indicated concentrations of the target DNA PCR products in the presence of a mixture of P-2 probes and Ampligase (see text for details). (B) An array of Msp T/C SNP P-1 oligomers hybridized with 100 fmol of target DNA from three individuals with AA, GG, and AG genotypes.
Further reduction in spot size (to 1 nl per spot) reduced the lower limit of target DNA detection to ≈5–10 fmol (data not shown). Nevertheless, 1-nl volumes of P-1 probes can be spotted at high density with excellent strength signals and allele discrimination provided that each target DNA is hybridized at a concentration of 100 fmol per 100-μl reaction. This is exemplified by the data shown in Fig. 3B. Here 1 nl of P-1 probes (T vs. C) for the MspHOXB locus was spotted from different concentration stock solutions by using a microarray contact printer. For each of the five P-1-T and P-1-C probe concentrations, a total of 60 1-nl spots were arrayed in three rows of 20, a total of 600 spots per 36-mm2 chip. Chips were hybridized with 100 fmol of target DNA (PCR product from the Msp locus of individuals with AA, GG, or AG genotypes, respectively) in the presence of P-2 probe and Ampligase. This target DNA concentration corresponds to ≤1 μl of a standard PCR. Here strong signals are seen at all P-1 probe concentrations, and SNP typing was straightforward. However, when signals from AA and GG homozygous target DNAs are compared, a higher level of nonspecific background was obtained when the mismatched base pair was a G/T pair. Discrimination of G/C from G/T is a common problem in both primer extension and ligation-based SNP assays.
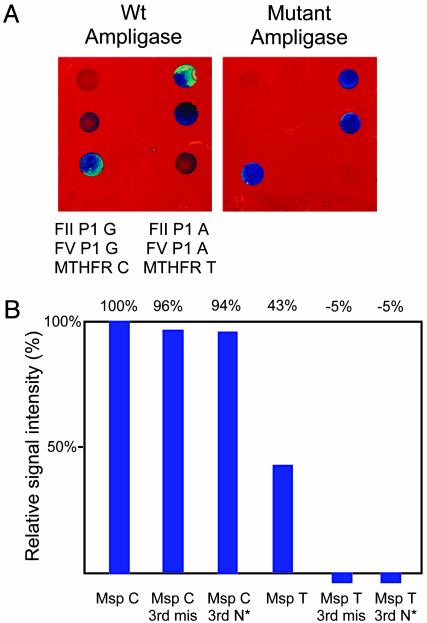
To address this background problem we investigated the inclusion of an additional mismatched base or a nitropyrrole nucleotide 3 nt from the 3′ terminus of the P-1 probe. Such changes are known to destabilize the mismatched probe-target hybrid further (20). In addition, the Lys-294 → Arg mutant of T. thermophilus DNA ligase, reported to have a 10- to 100-fold improvement in ligation of perfectly matched hybrids relative to mismatched hybrids (19), was cloned, purified, and used in place of wild-type enzyme. As shown in Fig. 4, both methods markedly decreased nonspecific signal from mismatched probe–target DNA duplexes. Because the production of mutant ligase is easier and less expensive than the synthesis of third base-mismatch probes, the mutant ligase has been incorporated into the standard assay for SNP typing of PCR amplicon DNA targets (see Methods and Materials).
Fig. 4.
Increased discrimination of G/A and C/T mismatches using a mutant T. thermophilus DNA ligase (A) or a nitropyrrole base (N*) or a second mismatched base 3 nt from the 3′ terminus of the P-1 probe (B). (A) P-1 probes for Factor V (FV), Factor II (FII), and MTHFR gene mutations (G-A, G-A, and C-T, respectively) hybridized to a mixture of PCR products in the presence of either the wild type (Wt) or the K294 mutant T. thermophilus ligase. (B) Relative signal intensity from Msp C/T SNP P-1 probes with or without a third base-mismatch (3rd mis) or nitropyrrole substitution.
Analysis of SNPs Associated with Increased Risk of Venous Thromboembolism (VTE). The fidelity of the biosensor SNP assay was evaluated by using sample material previously genotyped by using restriction fragment length polymorphism (RFLP) analysis and either the Invader or TaqMan systems.
VTE is a common medical problem with high morbidity. There are 600,000 cases of pulmonary embolism or deep VTE a year in the USA, of which only one-third are clinically identified. Certain genetic polymorphisms increase risk for VTE; these include the Factor VLeiden variant (a Gly-1691 → Ala change that results in an Arg-506 → Gln amino acid substitution), the prothrombin (Factor II) Gly-20210 → Ala variant that causes a transition in the 3′ untranslated region of the Factor II mRNA, and the homocysteine methylene tetrahydrofolate (MTHFR) gene Cys-677 → Thr transition that results in an Ala-222 → Val protein change. Heterozygous Factor VLeiden, for example, increases the risk of VTE 2.8-fold, and heterozygous prothrombin Gly-20210 → Ala increases the risk 1.8-fold in a group of young males (21, 22). Inheritance of both of these mutations, however, further increases the VTE risk to 16.5-fold (21). There are numerous other polymorphisms in genes associated with blood coagulation; however, most have never been evaluated for their role in the pathogenesis of VTE. To test the potential utility of biosensor chips as a platform for analyzing a large panel of SNPs that potentially could contribute to the overall risk factor for VTE, we constructed a hand-spotted test chip with allele-discrimination oligonucleotides for Factor VLeiden, MTHFR*C667T, and Factor II Gly-20210 → Ala.
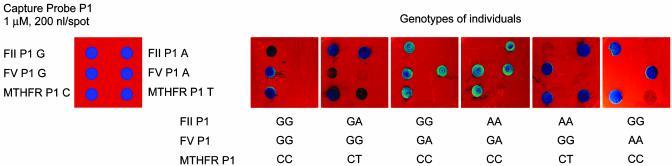
DNA samples from 100 individuals at risk for VTE were analyzed. These samples had been assayed previously for the Factor V, Factor II, and MTHFR mutations in the Marlar Laboratory by using the Third Wave Invader assay system. We were blind to these results. PCR primers for the SNP loci were synthesized, and conditions for PCR amplification of all three loci simultaneously were established (see Table 3). PCR products from each individual were assayed on biosensor chips under the standard assay conditions by using wild-type Ampligase. Typical results are shown in Fig. 5. When the sample identities were decoded, the data collected on the biosensor chip were 100% concordant with SNP typing generated by the Invader assay.
Fig. 5.
Detection of point mutations in Factor II (FII), Factor V (FV), and MTHFR genes associated with increased risk for VTE. Wild-type alleles are on the left side of the chips. Genotype data from six individuals are shown.
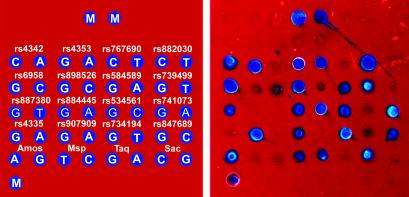
SNP Genotyping of Chromosome 17 Markers in an African-American Population. The utility of the biosensor chip for SNP typing in a population-genetics study was tested by arraying P-1 capture probes for 20 different SNP sites mapping to human chromosome 17 and then hybridizing these probes with DNA from 50 individuals in an African-American population. Three of these markers (the HOXB4 Msp rs3809782, HOXB3 Taq rs4646992, and HOXB6 Sac rs4646993) had been typed previously in the Kidd Laboratory by using RFLP analysis, whereas a fourth marker (rs4353 in the ACE gene) had been typed by using TaqMan assays. PCR products from each individual were prepared, pooled, and analyzed on the biosensor chips as described in Methods and Materials. A representative image is shown in Fig. 6.
Fig. 6.
Genotyping of 20 SNP markers in an African-American population. (Left) SNP designation and SNP nucleotides are listed in template array. M-orientation markers, covalently bound deoxyadenosine 20 with 3′-terminal biotin. (Right) Chip image for a single individual.
The typings obtained by using the biosensor chip were compared with the data for the four loci analyzed previously. There was complete concordance between the typings at the rs4353 locus (TaqMan), but there were a total of eight discrepancies at the three HOXB loci (RFLP). Reanalysis of these sites indicated that all discrepancies resulted from typing miscalls in the original RFLP data set. The data from all loci were also tested for deviation from Hardy–Weinberg expectation by using hwsim, a Monte Carlo permutation procedure for small sample sizes (http://info.med.yale.edu/genetics/kkidd/programs.html). By a conservative one-tailed test, none of the 20 loci deviated significantly (P < 0.01, uncorrected for multiple tests) from Hardy–Weinberg ratios. The allele frequencies observed at all loci are given in Table 4, which is published as supporting information on the PNAS web site.
In summary, the 100% concordance with typing data generated by TaqMan and Invader SNP assays and the ability to identify errors in the less rigorous RFLP technique indicate that SNP typing on biosensor chips is highly accurate.
Discussion
We have described a ligation-based, optical-detection platform for SNP genotyping that is simple, rapid, and robust, can be multiplexed extensively, and has a lower limit of target-detection sensitivity between 5 fmol and 100 amol (similar to fluorescencedetection methods) in an assay that takes <45 min. Spot density has a large effect on the limit of detection that is simply related to the thermodynamics of hybridization to surface-bound oligonucleotides: As the spot density is increased, the total amount of probe available to react is reduced. This can be overcome to some degree by raising the input concentration of the spotted probe, but eventually hybridization efficiency will be reduced as the packing density increases above an optimum.
One of the strengths of a ligation-based assay is that the design of SNP probes is very straightforward and involves no significant modeling or probe testing beyond that required to ascertain the quality of probe synthesis and to rule out the inclusion of repetitive sequence elements or palindromic structures. Indeed, each P-1 and P-2 probe for all loci tested (40/40 sets correctly synthesized) worked well under the standard assay condition without the requirement for optimization. Having the 5′ end of the 40-mer P-1 capture probe anchored to the chip ensures that any probe molecules that are prematurely terminated during synthesis do not interfere with downstream ligation reactions. Thus, probes can be used routinely without purification. These characteristics significantly reduce the cost of developing new SNP probe sets.
By formatting the ligation assay on the chip surface, analysis is simplified. The chip dimension allows for placement of chips into 96-well plates, which can be processed by using commercial pipetting robots, thus increasing overall throughput. Because spotting density of >300 per chip is readily obtainable, ≥15,000 SNP typings can be done by using a single microtiter plate of chips. The chips can be read by a simple charge-coupled device camera and alleles can by discriminated spatially, allowing for inexpensive and highly accurate genotype determinations. Alternatively, the assay is simple and robust enough to be used as a “fieldable assay” for low-density (<20 SNPs per chip) genotype tests with visual readout.
The assay as currently formatted uses PCR amplicons as the target DNA; thus, it has limitations similar to other procedures that incorporate PCR amplification. However, one of the intriguing features of the biosensor is the potential to push the current detection sensitivity to extremely high (subattomole) levels. The significance of this would be the possibility of performing SNP assays directly on genomic DNA with or without the application of whole-genome amplification techniques such as multiple displacement amplification (23, 24) that introduce little if any genetic bias during the amplification process. A number of strategies can be envisioned to enhance the overall sensitivity of target DNA detection further. The simplest and most straightforward strategy is to maintain the existing assay structure but introduce multiple biotins onto each P-2 probe. This should increase the number of anti-biotin IgG-HRP conjugates recruited by each P-1–P-2 ligation event and decrease the number of hybridizing target DNA molecules required to produce the prerequisite increase in thin-film thickness to generate a detectable surface-color change. Indeed, preliminary experiments (X.Z., H. Chen, and D.C.W., unpublished data) demonstrate a 50- to 100-fold increase in assay sensitivity with biotinylated probes decorated with an avidin-biotinylated dextran copolymer or P-2 probes with a 3′-terminal sequence, complementary to a small circular DNA template, that can be extended by rolling-circle amplification (25, 26) in the presence of biotin-dUTP. Although a ≥1,000-fold increase in detection sensitivity will likely be required before total human genomic DNA can be used for SNP typing on biosensor chips without prior site-specific PCR amplification, analysis of eukaryotic genomes with less genetic complexity (e.g., Drosophila, Arabidopsis, or Caenorhabditis elegans) should now be feasible.
Supplementary Material
Acknowledgments
We thank Dr. Efim Golub for preparing the mutant Ampligase; Drs. Kevin White and Tong-rei Li for helping us to spot P-1 probes with their microarray printer; and Drs. Patricia Bray-Ward and Barry Polisky for thoughtful discussions. This work was supported by Department of Energy Grant DE-FG02-00ER63058 (to D.C.W.), National Institutes of Health Grant P01GM57672 (to K.K.K.), and a grant from the Department of Veterans Affairs (to R.A.M.).
Abbreviations: SNP, single-nucleotide polymorphism; HRP, horseradish peroxidase; VTE, venous thromboembolism; RFLP, restriction fragment length polymorphism; MTHFR, methylene tetrahydrofolate.
References
- 1.Kirk, B. W., Feinsod, M., Favis, R., Kliman, R. M. & Barany, F. (2002) Nucleic Acids Res. 30, 3295–3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.International SNP Map Work Group (2001) Nature 409, 928–933.11237013 [Google Scholar]
- 3.Halushka, M. K., Fan, J. B., Bentley, K., Hsie, L., Shen, N., Weder, A., Cooper, R., Lipshutz, R. & Chakravarti, A. (1999) Nat. Genet. 22, 239–247. [DOI] [PubMed] [Google Scholar]
- 4.Nordborg, M. & Tavare, S. (2002) Trends Genet. 18, 83–90. [DOI] [PubMed] [Google Scholar]
- 5.Syvanen, A. C. (2001) Nat. Rev. Genet. 2, 930–942. [DOI] [PubMed] [Google Scholar]
- 6.Kwok, P. Y. (2001) Annu. Rev. Genomics Hum. Genet. 2, 235–258. [DOI] [PubMed] [Google Scholar]
- 7.Kan, Y. W., Lee, K. Y., Furbetta, M., Angius, A. & Cao, A. (1980) N. Engl. J. Med. 302, 185–188. [DOI] [PubMed] [Google Scholar]
- 8.Marras, S. A., Kramer, F. R. & Tyagi, S. (2003) Methods Mol. Biol. 212, 111–128. [DOI] [PubMed] [Google Scholar]
- 9.Pastinen, T., Raitio, M., Lindroos, K., Tainola, P., Peltonen, L. & Syvanen, A. C. (2000) Genome Res. 10, 1031–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hsu, T. M., Law, S. M., Duan, S., Neri, B. P. & Kwok, P. Y. (2001) Clin. Chem. (Washington, D.C.) 47, 1373–1377. [PubMed] [Google Scholar]
- 11.Pickering, J., Bamford, A., Godbole, V., Briggs, J., Scozzafava, G., Roe, P., Wheeler, C., Ghouse, R. & Cuss, S. (2002) Nucleic Acids Res. 30, e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ross, P., Hall, L., Smirnov, I. & Haff, L. (1998) Nat. Biotechnol. 16, 1347–1351. [DOI] [PubMed] [Google Scholar]
- 13.Ronaghi, M. (2001) Genome Res. 11, 3–11. [DOI] [PubMed] [Google Scholar]
- 14.Hall, J. G., Eis, P. S., Law, S. M., Reyanldo, L. P. & Prudent, J. R. (2001) Proc. Natl. Acad. Sci. USA 97, 8272–8277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Livak, K. J. (1999) Genet. Anal. 14, 143–149. [DOI] [PubMed] [Google Scholar]
- 16.Sandstrom, T., Steinberg, M. & Nygren, H. (1985) Appl. Opt. 24, 472–479. [DOI] [PubMed] [Google Scholar]
- 17.Jenison, R., La, H., Haeberli, A., Ostroff, R. & Polisky, B. (2001) Clin. Chem. (Washington, D.C.) 47, 1894–1900. [PubMed] [Google Scholar]
- 18.Jenison, R., Yang, S., Haeberli, A. & Polisky, B. (2001) Nat. Biotechnol. 19, 62–65. [DOI] [PubMed] [Google Scholar]
- 19.Luo, J., Bergstrom, D. E. & Barany, F. (1996) Nucleic Acids Res. 24, 3071–3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo, Z., Liu, Q. H. & Smith, L. M. (1997) Nat. Biotechnol. 15, 331–335. [DOI] [PubMed] [Google Scholar]
- 21.Ridker, P. M., Hennekens, C. H., Selhub, J., Miletich, J. P., Malinow, M. R. & Stampfer, M. J. (1997) Circulation 95, 1777–1782. [DOI] [PubMed] [Google Scholar]
- 22.Rosendaal, F. R. (1997) Semin. Hematol. 34, 171–187. [PubMed] [Google Scholar]
- 23.Dean, F. B., Hosono, S., Fang, L., Wu, X., Faruqi, A. F., Bray-Ward, P., Xun, Z., Zong, Q., Du, Y., Du, J., et. al. (2002) Proc. Natl. Acad. Sci. USA 99, 5261–5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lage, J. M., Leamon, J. H., Pejovic, T., Hamann, S., Lacey, M., Dillon, D., Segraves, R., Vossbrinck, B., Gonzalez, A., Pinkel, D., et. al. (2003) Genome Res. 13, 294–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lizardi, P. M., Huang, X., Zhu, Z., Bray-Ward, P., Thomas, D. C. & Ward, D. C. (1998) Nat. Genet. 19, 225–232. [DOI] [PubMed] [Google Scholar]
- 26.Zhong, X. B., Lizardi, P. M., Huang, X., Bray-Ward, P. & Ward, D. C. (2001) Proc. Natl. Acad. Sci. USA 98, 3940–3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.