Abstract
To understand the origin and emergence of pathogenic bacteria, knowledge of the genetic inventory from their nonpathogenic relatives is a prerequisite. Therefore, the 2.11-megabase genome sequence of Wolinella succinogenes, which is closely related to the pathogenic bacteria Helicobacter pylori and Campylobacter jejuni, was determined. Despite being considered nonpathogenic to its bovine host, W. succinogenes holds an extensive repertoire of genes homologous to known bacterial virulence factors. Many of these genes have been acquired by lateral gene transfer, because part of the virulence plasmid pVir and an N-linked glycosylation gene cluster were found to be syntenic between C. jejuni and genomic islands of W. succinogenes. In contrast to other host-adapted bacteria, W. succinogenes does harbor the highest density of bacterial sensor kinases found in any bacterial genome to date, together with an elaborate signaling circuitry of the GGDEF family of proteins. Because the analysis of the W. succinogenes genome also revealed genes related to soil- and plant-associated bacteria such as the nif genes, W. succinogenes may represent a member of the epsilon proteobacteria with a life cycle outside its host.
Keywords: Helicobacter, Campylobacter, epsilon proteobacteria, bacterial pathogenicity
Species of the group of Campylobacteraceae and Helicobacteraceae are important pathogens in humans and animals (1, 2). Only recently Campylobacter jejuni was recognized as the causative agent of serious illnesses such as Guillain–Barré syndrome (3) and human gastroenteritis (4). The most prominent representative of the Helicobacteraceae is Helicobacter pylori, which has been demonstrated to cause ulcers and gastric cancer in humans (3). These organisms belong together with Wolinella succinogenes to the epsilon subclass of the proteobacteria, as has been demonstrated by 16S rRNA analysis (5, 6). Despite being a member of the Helicobacteraceae, W. succinogenes was shown to be phylogenetically intermediate between both families (6). W. succinogenes was originally isolated from the rumen of cattle (7) and has since been reisolated and typed successfully by molecular methods (8). Currently there is no evidence of W. succinogenes having adverse effects on the health of humans or animals. For these reasons, W. succinogenes has been considered to be a nonpathogenic, host-associated organism.
W. succinogenes is a nonfermenting bacterium that grows by different modes of anaerobic respiration (8–10). Furthermore, it has been reported to grow in the presence of 2% oxygen (7, 11). The darting motility of W. succinogenes has triggered several projects that investigated the unique aspects of its monotrichous flagellation and the insertion of the flagellar motor into the pole of the cell (12–15).
The completion of the W. succinogenes genome yields unexpected findings in regard to gene content that were previously unknown for host-associated bacteria.
Materials and Methods
Genome Sequencing. Genomic DNA from W. succinogenes DSMZ 1740 was isolated from cells grown in liquid cultures by using the Qiagen genome DNA kit (Qiagen, Hilden, Germany). DNA libraries with insert sizes of 1–2 kb, 3–5 kb (TOPO Shotgun subcloning kit, Invitrogen), and 40 kb (Epicentre Technologies, Madison, WI) were constructed and end-sequenced to 8-fold coverage (16). Remaining gaps were closed by direct sequencing with chromosomal DNA as a template. The final sequencing error rate was estimated to be <0.67 × 10–5 by using the phred/phrap/consed software package (17–20). The W. succinogenes genome sequence has been assigned EMBL accession no. BX571656 and is also available at www.wolinella.mpg.de.
Annotation and Analysis. Curation and annotation of the genome was done by using the annotation package gendb (21). ORFs were predicted by the program glimmer (22), which is integrated into the gendb package. Annotation of the identified ORFs was accomplished on the basis of similarity searches against different databases and manual curation. Similarity searches were performed by using blastx (23) against the nonredundant database on protein level. After the blast procedure all ORFs being shorter than 150 bp and having no blast hit with an E value <10–15 were discarded. The remaining ORFs were postprocessed by using the program rbsfinder (24) to correct the start codons by searching for ribosomal binding sites. Finally, each putatively identified gene was assigned to a category of the Clusters of Orthologous Groups (COG) database (25).
Genomic Comparison. The C. jejuni (GenBank accession no. AL111168), H. pylori 26695 (GenBank accession no. AE000511), and J99 (GenBank accession no. AE001439) genomes were downloaded. Homology searches were conducted against the genomes and plasmid sequences of H. pylori (26, 27) and C. jejuni (28) on the nucleotide and amino acid level by using the blast software package (23). The blast output was processed further to determine syntenic regions between two genomes.
Results
Genome Features. The genome of W. succinogenes consists of a circular chromosome of 2,110,355 bp with an average GC content of 48.5% (Table 1). No plasmids were found during the genome analysis, nor were any described in literature. The origin of replication (ori) is clearly detectable by a bias of G toward the leading strand (GC skew) (29). Because the dnaA gene has been found to localize in this region, its start codon has been designated as the zero point of the genome. The genome shows a high gene density with 2,046 predicted ORFs and a coding area of 94.0%. For 1,260 ORFs (61.5%) a putative function could be assigned, 297 ORFs (14.5%) have matches to hypothetical proteins, and 490 ORFs (23.9%) do not have a database match. Three rRNA operons consisting of 16S, 23S, and 5S rRNA genes were found, together with 40 tRNAs representing all 20 amino acids (Table 1).
Table 1. Genome features.
| Species | W. succinogenes |
|---|---|
| Strain | DSMZ 1740 |
| Size, bp | 2,110,355 |
| G + C content, % | 48.5 |
| ORFs | |
| Predicted number of ORFs | 2,046 |
| % of genome coding | 94.0 |
| Average length, bp | 964 |
| % ATG initiation codons | 70.2 |
| % GTG initiation codons | 14.8 |
| % TTG initiation codons | 15.0 |
| % other initiation codons | n.d. |
| IS elements | |
| Complete IS1302 copies | 12 |
| Partial IS1302 copies | 1 |
| Complete ISWsu1203 copies | 1 |
| Partial ISWsu1203 copies | 3 |
| RNA elements | |
| 23S-5S rRNA | 3 |
| 16S rRNA | 3 |
| tRNAs | 40 |
n.d., not determined.
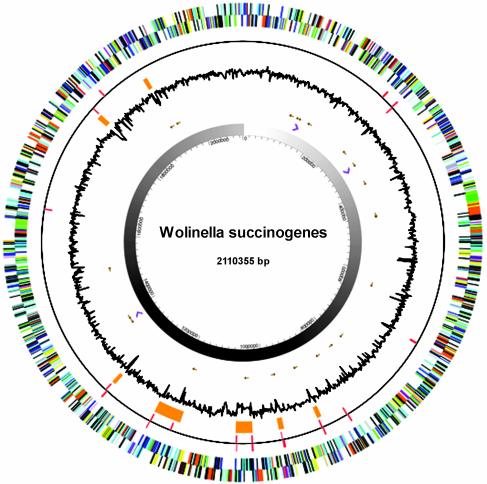
Besides the three rRNA clusters, additional regions (>5 kb) with repeated sequences were identified. One such region contains two formate dehydrogenase operons, each consisting of five genes (fdhE, -A, -B, -C, and -D) but with opposing transcriptional direction (30). Furthermore, an additional fdhE, -A, -B, and -C locus was found together with a third fdhD gene that apparently has been transposed to a location unrelated to its neighboring genes. Other regions with repetitive sequences are associated with families of paralogous proteins, which comprise ≈21% of all proteins. The majority of proteins falling into this category encompass ATP-binding cassette transporters (gene copy no. 51), histidine kinases (no. 39), response regulators (no. 52), methyl-accepting chemotaxis proteins (no. 31), GGDEF-family proteins (no. 28), transposases (no. 17), conserved hypothetical proteins (no. 20), and hypothetical proteins (no. 39). Several regions with a large deviation in GC content are observable, often flanked by insertion elements (ISs) on opposing strands (Fig. 1). Twelve of these previously described IS1302-type ISs (31) are present in the genome, together with four copies of a previously undescribed multicopy element termed ISWsu1203.
Fig. 1.
Circular representation of the W. succinogenes genome. The outer-most circles show predicted protein-coding regions on the + (wheel 1) and – (wheel 2) strands. The red bars on wheels 3 and 4 indicate ISs of the type IS1302. They are found on both strands, frequently flanking genomic islands and islets. These are depicted by orange bars on wheel 5 and are collinear with regions that show a large deviation from the average GC content (wheel 6). Non-protein-coding genes such as tRNA and rRNA genes are shown as brown and purple arrows on wheels 7 and 8. The origin of replication was defined by a bias of G over C (GC skew), which is clearly observable in wheel 9.
The average GC content drops in the flanked areas from 48.5% to ≈36% and probably arose from DNA uptake, mediated either by natural transformation or phage-mediated transduction. The W. succinogenes genome shows little organization of genes that are functionally interconnected and no evidence for extensive transcriptional units other than conserved operon structures encoding multisubunit enzymes involved in primary metabolism. Genes coding for complex cellular structures such as the flagellar motor are found at numerous different positions in the genome. This disorganization may have arisen from high rates of recent recombination, because no genome-wide collinearity is detectable to the genomes of H. pylori and C. jejuni.
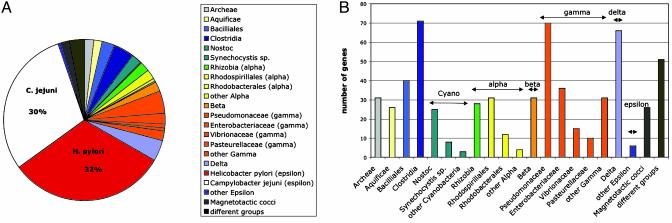
Phylogeny. W. succinogenes is a member of the epsilon proteobacteria. Based on 16S rRNA phylogeny, W. succinogenes was shown to be more closely related to H. pylori than to C. jejuni, a fact that has led to its classification as a member of the Helicobacteraceae (6). Comparison of the complete predicted set of W. succinogenes proteins to the NCBI nonredundant database (cutoff 10–15) results in an almost even distribution of high-scoring pairs between H. pylori (32%) and C. jejuni (30%) (Fig. 2A). A total of 614 proteins was found to show highest homology between W. succinogenes and C. jejuni, whereas 655 proteins have their closest hit in H. pylori. The remaining 38% of high-scoring pairs are distributed between a wide variety of other taxa, such as gamma proteobacteria (no. 162), Clostridia (no. 71), alpha proteobacteria (no. 71), delta proteobacteria (no. 71), and Cyanobacteria (no. 40) (Fig. 2B). Because the W. succinogenes genome is 25% larger than its two epsilon-proteobacteria relatives, the additional genetic information might have either originated from their last common ancestor or been acquired via horizontal gene transfer. The genomic islands with deviating GC content that are described below suggest that the latter possibility accounts for a substantial amount of the W. succinogenes genetic information.
Fig. 2.
Phylogenetic analysis and taxonomy blast of the W. succinogenes genes. Each of the 2,046 W. succinogenes ORFs except the rRNAs were blasted on the protein level (blastp) against all NCBI database entries. (A) Taxonomic distribution of the genes. Approximately 32% of the predicted W. succinogenes proteins do have their closest homologue in H. pylori, another third in C. jejuni, and the remaining 38% in other organisms. Prominent herein are the Cyanobacteria (see also Fig. 3C) and the Enterobacteriaceae as well as the Pseudomonaceae (B).
Metabolism. Central metabolic routes. Cells of W. succinogenes use fumarate as the sole carbon source during growth by anaerobic fumarate respiration in a minimal medium. The biosynthetic pathways for conversion of fumarate were predicted previously (32) and have now been confirmed by the genome sequence (see Fig. 4, which is published as supporting information on the PNAS web site, www.pnas.org). Complete biosynthetic pathways for the formation of amino acids, purine and pyrimidine nucleotides, as well as fatty acids and phospholipids from intermediates of the central metabolism could be established. The same holds true for the biosynthesis of various enzyme cofactors and prosthetic groups such as NAD(P)+, thiamine pyrophosphate, pyridoxal phosphate, riboflavin, heme, molybdopterin, folate, pantothenate, or biotin. W. succinogenes does not ferment carbohydrates such as glucose, and neither glucokinase nor a glucose transport system were detected. Surprisingly, a phosphofructokinase-encoding gene (WS1028) was found, indicating a complete glycolytic pathway from glucose 6-phosphate to pyruvate. The lack of a glucose transport system is not compensated by alternative transporters such as a hexose phosphate transporter. Therefore, it is likely that W. succinogenes uses its glycolytic enzymes solely for gluconeogenesis. Key enzymes of the Entner–Doudoroff pathway, the oxidative pentose phosphate pathway, and the glyoxylate cycle as well as orthologues of phosphotransacetylase and acetate kinase have not been found.
Energy metabolism. In addition to previously identified genes coding for electron transport enzymes involved in anaerobic respiration (8–10), six thus-far-unknown molybdenum-containing oxidoreductase complexes were predicted. One of these enzymes is probably a selenocysteine-containing formate dehydrogenase (WS0733), whereas other enzymes may play a role in reduction of dimethyl sulfoxide or trimethylamine N-oxide.
W. succinogenes contains genes encoding putative enzyme complexes typical of aerobic respiration that closely resemble those of the microaerobically growing species C. jejuni and H. pylori (33). These complexes comprise NADH:quinone oxidoreductase, cytochrome bc1 complex, and two distinct terminal oxidases (cytochromes cbb3 and bd). Furthermore, a putative succinate:quinone reductase that contains an unusual membrane anchor subunit typically found in several Archaea is present (34). No candidate for a potential catalase gene was found. However, several protective enzymes that may cope with reactive oxygen species such as superoxide and peroxide are encoded in the genome.
Transport processes. The uptake and exchange of the dicarboxylates fumarate and succinate are essential with respect to fumarate respiration of W. succinogenes (35). In addition to the two previously described sodium-dependent dicarboxylate antiporters DcuA and DcuB and a putative dicarboxylate uptake system that belongs to the tripartite ATP-independent periplasmic transporters, the genome is predicted to contain a wealth of inner and outer membrane proteins that are likely to be involved in transport of various cations, anions, amino acids, and oligopeptides (Fig. 4). The majority of these are ATP-binding cassette transporters, permeases of the major facilitator superfamily, or drug/metabolite transporter superfamily. However, no phosphotransferase (group-translocation) system has been identified.
As has been reported for the genomes of H. pylori and C. jejuni, a large variety of different uptake systems for ferrous or ferric iron (36, 37) is also found in W. succinogenes. Two distinct feoAB loci (WS0955/WS0956 and WS1412/WS1413) may encode ferrous iron uptake systems located in the cytoplasmic membrane. An ATP-binding cassette transporter system for ferric iron transport of the Fec type is also present (WS1123–WS1126). Furthermore, a total of 11 putative outer membrane proteins (COG description CirA) that may play a role in TonB-dependent uptake of ferric iron compounds were found. There are three sets of exbBD genes in the W. succinogenes genome. Two regulator proteins of the Fur type (WS0545 and WS2043) that are involved in expression of iron uptake or oxidative stress-defense systems in other organisms are present (37).
Genomic Islands and Virulence Factor Homologues. Mobile genetic elements. The W. succinogenes genome contains two distinct multicopy elements belonging to the family of ISs: 12 identical full-length copies of the IS1302 insertion sequence (31) and a single variant. No single nucleotide polymorphisms are detectable among the IS1302 elements even in their noncoding regions, suggesting that the entire IS1302 sequence is either a recent acquisition (or under selective pressure) or is being maintained by homogenization via gene conversion. The predicted coding region of the transposase, together with two detectable domains is indicative for functional IS3-type elements (38). The various integrations of the IS1302 element lead to disruption of eight protein-coding genes in the sequenced strain DSMZ 1740. In addition to the IS1302, one full-length copy of a formerly undiscovered IS, named ISWsu1203, was found together with three copies that were disrupted by an insertion of IS1302. The insertion of an IS into another preexisting one may be explained by an insertion mechanism that is generally biased toward AT-rich stretches of sequence (39). In addition to the described ISs, W. succinogenes harbors two integrases, which are likely to be of bacteriophagal origin.
Genomic islets and islands. The W. succinogenes genome contains several distinct regions that can be categorized as “flexible genomic islands and islets” (40) with sizes ranging from 2 up to 45 kb. These genomic islands and islets are congruent with the AT-rich regions depicted in Fig. 1 (wheels 4 and 5) and deviate from the average GC content of the core genome by as much as 12.5%. This fact together with the observed colocalization of the genomic islands and islets with tRNA loci and the presence of mobile genetic elements (ISs) flanking these sites strongly suggest the recent transfer of genetic material into W. succinogenes (41, 42).
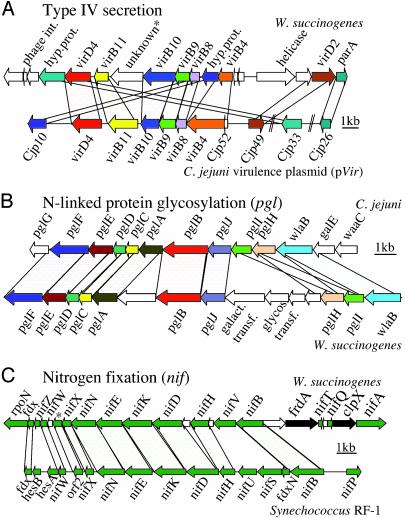
This horizontal gene transfer is particularly evident for the genomic island I with a size of ≈28 kb and a GC content of 39%, which encodes a cluster of 23 genes (Fig. 3A). Remarkably, the central section of the genomic island I reveals an almost undisturbed synteny with one third of the C. jejuni virulence plasmid pVir (Fig. 3A) (43). Ten of the 23 genes are orthologues of components assembling the type IV secretion apparatus, which is also found in H. pylori and functions as a delivery system of biopolymers into a host cell (44). The type IV secretion gene cluster from the W. succinogenes genomic island I and the Campylobacter virulence plasmid pVir, however, do not exhibit the gene order found in the pathogenicity-associated island (cagPAI) of H. pylori, suggesting a different origin of the H. pylori genomic island (45). Of the gene products necessary for the assembly of a minimal core structure of the type IV secretory machinery, VirB4, VirB9, VirB10, VirB11, and VirD4 are present, arguing for a functional type IV secretion machinery in W. succinogenes (46). Furthermore, the W. succinogenes island harbors 10 additional genes, five of which are interspersed in between the vir genes. Among them are a set of DNA-modifying enzymes such as the plasmid-partitioning gene (parA), a DNA helicase, and a gene with a conserved topoisomerase domain, all of which suggest a DNA-processing mechanism during transfer and integration.
Fig. 3.
Syntenic regions of the W. succinogenes genome. (A) Schematic representation of the W. succinogenes genomic island I and the syntenic region of the C. jejuni 81–176 virulence plasmid (pVir). Both loci encode multiple genes involved in type IV secretion. Connecting lines indicate corresponding orthologous genes with high homology on the protein level. (B) Schematic representation of the syntenic W. succinogenes and C. jejuni protein-glycosylation loci (pgl). (C) Schematic representation of the W. succinogenes nitrogen-fixation (nif) gene cluster in comparison to the partly syntenic nif cluster of the cyanobacterium Synechococcus RF-1. The interspersed white genes encode hypothetical proteins. The star indicates a W. succinogenes protein containing a DUF269 domain, also found within the nitrogen-fixation operons of Cyanobacteria.
Genomic island I is inserted in the W. succinogenes core genome via disruption of a tRNAmet locus. Such tRNA loci repeatedly served as target sites of phage-mediated transduction events due to their sequence homologies to attachment sites of bacteriophagal integrases (47). The two flanking IS1302 ISs suggest that this island may be mobilized as a complex transposon. A transcriptional regulator gene of araC type neighbors the island; this class of regulators is known to be involved primarily in regulating pathogenicity islands in other bacteria but is also present in nonpathogenic organisms (48).
Also for genomic islet I there is clear evidence of a horizontal acquisition mediated via a bacteriophagal transduction event. This 4.7-kb region is integrated into the W. succinogenes genome by disrupting a tRNAphe locus, as has been observed for genomic island I. Phage transfer of this region is supported by the finding of several ORFs and pseudogene fragments of bacteriophagal origin, including an integrase and an integrase fragment, as well as a GC content of 36% (49).
Pathogenicity-Related Genes Outside Genomic Islands. W. succinogenes shares a large number of genes that were identified in H. pylori and C. jejuni as virulence factors (28, 43, 46). These include several hemolysin-related genes, adhesion factors, and pili-generating proteins, invasins, antigenicity factors, resistance genes, and proteases as well as the neutrophil-activating protein and the virulence factor MviN (see Table 2, which is published as supporting information on the PNAS web site).
However, the most striking example is the secreted C. jejuni invasion antigen B (ciaB), which is a key pathogenicity factor of C. jejuni and is essential for invading the host cell in the infection process (50). Interestingly, thus far this protein is exclusively found in C. jejuni and W. succinogenes but not in any other organism. Despite the large number of orthologues shared by W. succinogenes and H. pylori, the pathogenicity-related genes rather resemble the pattern found in C. jejuni instead of the one of H. pylori. The fact that the nonpathogenic W. succinogenes groups large numbers of virulence-factor and fitness-gene homologues suggests that these genes might not only be involved in pathogenic responses toward a host but also might be involved in maintaining a symbiotic or commensal relationship with a host.
Protein Glycosylation. N-linked glycosylation is the most frequent protein-modification system in eukaryotes and can be found also in Archaea. Recently an N-linked glycosylation system was described in C. jejuni as the first example of such a system in bacteria (51, 52). Central to this system in C. jejuni is the pglB gene, which encodes a protein with high homology to Stt3p, an essential component of the N-linked protein-glycosylation system in eukaryotes. Interestingly, W. succinogenes is the second bacterial organism harboring a pglB homologue. The C. jejuni gene cluster of 16.7 kb was shown to contain a total of 13 genes involved in synthesis, transport, and transfer of glycosyl residues onto at least 38 target proteins (53). Despite the fact that the W. succinogenes genome in general has not maintained a syntenic order of its transcriptional units in comparison to C. jejuni, the glycosylation cluster is one of the few examples where this is the case. The W. succinogenes N-glycosylation cluster of 18.5 kb has maintained its order relative to the origin of replication when compared with C. jejuni (Fig. 3B). For 10 of the 13 genes the order is conserved in W. succinogenes, with two genes (pglH and pglI) having exchanged their positions. Homologues for the remaining three genes (pglG, galE, and waaC) are found separated from the cluster at isolated positions throughout the genome. In addition, five genes are found interspersed between pglA and pglB, as well as pglJ and pglH. The latter insertion harbors four genes, two of which are also clearly related to protein glycosylation. The presence of these additional genes, a galactosyl transferase (WS0046) and a glycosyl transferase (WS0047), might be indicative for the formation of a polyglycoside oligomer that deviates in its structure from the heptasaccharide described for C. jejuni (51). The hypothesis that W. succinogenes conducts N-linked protein glycosylation is supported by the fact that 29 of 38 described target proteins have orthologues in the W. succinogenes genome.
Signal Transduction. Two-component signal transduction systems (TCSTs) are widely used by bacteria to monitor their immediate environment and to control cellular processes in response to perceived stimuli. Extensive two-component signaling networks have been considered a hallmark of environmental strains capable of dwelling in a wide variety of growth conditions and bacteria with complex life cycles (54). Host-adapted organisms, in contrast, have lost genes coding for the cellular signaling circuitry by a wide margin (46, 55), because they adapted to niches with constant conditions. Surprisingly, W. succinogenes, despite being a host-associated bacterium, has the largest ever reported number of TCST proteins of all completed bacterial and archaeal genomes when normalized by genome size. Eighty genes coding for histidine kinases or their cognate regulators are found, resulting in a density of ≈38 TCST genes per megabase. Anabena sp., the previous leader in this category, has a total number of 195 genes within a 7.2-megabase genome (density ≈ 27 genes per megabase) (56). Domain analysis of the TCST genes reveals a total of 27 histidine kinases, 38 response-regulator genes, two histidine phospho-transfer proteins, and 13 hybrid histidine kinases, which consist of a kinase and a regulator domain on the same polypeptide. Response-regulator domains apparently have been duplicated most frequently, because a total of 54 of those domains have been found, resulting in genes with multiple response-regulator domains. The W. succinogenes TCST circuitry in general shows a wealth of combination with other signaling domains such as GAF, PP2C_SIG, PBPB, HDc, and PAS/PAC domains (57) that are highly reminiscent of the TCST genes found in lower eukaryotes such as Dictyostelium discoideum (58, 59). Noteworthy among these are four genes (WS0108, WS1021, WS0414, and WS0344) with a combination of response-regulator domains and two domains of unknown function (DUF1 and DUF2), which are also known as the GGDEF and EAL family of proteins (60). They may constitute an interface between the signaling networks of the two-component systems and the newly emerging signaling circuitry of the DUF gene products. In total, 26 DUF genes could be detected in the W. succinogenes genome, 13 of which harbor a single DUF1 domain, 7 with a DUF2 domain, and 6 genes that show a combination of both. Several of the DUF genes are found in the genome at neighboring positions in a similar arrangement as has been described for many of the TCST kinase-regulator pairs. Which cellular processes are controlled by the DUF genes is unclear at present, although a role in the regulation of the nif genes seems likely, because one of these genes (WS1386) and its homologues are found in the nif gene cluster of W. succinogenes and several other bacterial organisms.
The elaborate signaling and sensing capabilities of W. succinogenes are also shown in its chemotaxis system (54). The principal signaling modules consisting of the histidine kinase CheA, a linker protein CheW, and its regulator CheY are present only in single copies. No genes involved in the adaptation process such as CheR or CheB were found. In contrast, 34 paralogues of the chemotaxis receptor (methyl-accepting chemotaxis proteins) can be predicted. Additional paralogues coding for the linker proteins CheW and CheV were identified in two and four copies, respectively. All CheW and CheV are thought to be essential for integrating the signals coming from distinct subgroups of the 34 chemotaxis receptors, because all methyl-accepting chemotaxis proteins have to signal through a single CheA kinase.
This signal is subsequently forwarded by regulator CheY (WS0619) to the flagellar motor, which is ensembled largely by the same molecular components as described for H. pylori and C. jejuni (28, 61) and constitutes of at least 24 gene products.
Only few signaling genes were found that fall into categories other than the one described above. For example, a single gene was predicted as a putative adenylate cyclase (WS1633) together with three genes that categorize as protein phosphatases, two of which could function as phospho-tyrosine phosphatases (WS0702 and WS1323). Despite the obvious lack of eukaryotic-type kinases, these gene products could act on substrates that were phosphorylated by histidine kinases, as has been suggested (62).
Nitrogen-Fixation Genes. Numerous prokaryotes are capable of dinitrogen fixation, the conversion of nitrogen to ammonium. Diazotrophs can be found in a wide range of environments including soils, oceans, lakes, intestines, feces, and insects (63). Although nitrogen fixation (nif) has not been reported previously in epsilon proteobacteria, the annotation of the W. succinogenes genome resulted in the identification of 30 nif genes and related genes, which code for the structural subunits of nitrogenase, as well as for accessory proteins and regulators involved in activation and repression of the nif regulon (Fig. 3C). Of these genes, 23 are clustered between W. succinogenes genes WS1381 and WS1404, whereas seven others (WS0409, WS0410, WS0560, WS0836, WS2205, WS2206, and WS2213) were found at various locations, independent from the context of their neighboring genes. For the synthesis of the active nitrogenase enzyme complex ≈20 genes are required (64). Homologues of all genes essential for the transcription and regulation of a nitrogenase complex are present in W. succinogenes. The nifH, -D, and -K genes (WS1391, WS1392, and WS1394) encode the subunits of nitrogenase, whereas the products of nifE, -N, and -B genes (WS1388, WS1390, and WS1397) are required for the synthesis of the iron–molybdenum cofactor. The gene nifA (WS1404) is believed to act as a regulator of the nif regulon (65), together with the RNA polymerase sigma factor σ54, rpoN (WS1381), which is found downstream of the gene cluster.
A significant fraction of genes within the W. succinogenes nif gene cluster is taxonomically related to genes of Cyanobacteria as well as to the N2-fixing legume symbionts of the Rhizobiaceae family. This finding, together with the fact that these genes are syntenic to the nif gene cluster of Cyanobacteria, again raises the possibility of an acquisition from a mobile genetic element via horizontal gene transfer (Fig. 3C) (56, 66). Cyanobacteria or Rhizobiaceae enter the rumen of cattle by uptake of either eutrophic water or leguminose plants together with the attached rhizosphere containing Rhizobiaceae. The bovine rumen is known to serve as a habitat for a wide variety of microbial species from many taxa, allowing them to interact and exchange genetic information. The hypothesis of lateral gene transfer between W. succinogenes and Cyanobacteria/Rhizobiaceae is supported by the fact that the transposable elements (ISs) found in W. succinogenes show their highest homology to sequences originating from both groups of Cyanobacteria and Rhizobiaceae as well.
Conclusions
The sequencing and annotation of the W. succinogenes genome has revealed a genomic inventory that is highly similar to those of C. jejuni and H. pylori. Particularly the finding of syntenic genes from the Campylobacter virulence plasmid pVir, as well as the presence of the second only bacterial N-linked glycosylation gene cluster, revealed an unforeseen relationship between W. succinogenes and C. jejuni. Both organisms also share a similar set of metabolic capabilities while omitting features specific to H. pylori. The 29% larger genome of W. succinogenes, however, codes for genes not found in any of its epsilon-proteobacterial relatives such as genes for nitrogen fixation, extensive signaling capabilities, and an unusually complete set of metabolic pathways. This wealth of genetic information, which is clearly not required for host adaptation, contradicts the general paradigm of degrading genomes that has been observed with many host-associated organisms such as pathogenic or symbiotic bacteria. It therefore seems likely that W. succinogenes may not be restricted to its ecological niche in the bovine rumen.
Supplementary Material
Acknowledgments
We commemorate and thank Professor Achim Kröger, who passed away during the collaborational effort, for annotating the W. succinogenes metabolism. We thank G. Velicer for helpful discussions and critical comments on the manuscript. We are grateful to N. E. Wittekindt and S. Rendulic for critical reading of the manuscript.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: IS, insertion element; TCST, two-component signal transduction system.
Data deposition: The sequence reported in this paper has been deposited in the EMBL database (accession no. BX571656).
References
- 1.Park, S. F. (2002) Int. J. Food Microbiol. 74, 177–188. [DOI] [PubMed] [Google Scholar]
- 2.Solnick, J. V. & Schauer, D. B. (2001) Clin. Microbiol. Rev. 14, 59–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chowdhury, D. & Arora, A. (2001) Acta Neurol. Scand. 103, 267–277. [DOI] [PubMed] [Google Scholar]
- 4.Wassenaar, T. M. & Blaser, M. J. (1999) Microbes Infect. 1, 1023–1033. [DOI] [PubMed] [Google Scholar]
- 5.Paster, B. J. & Dewhirst, F. E. (1988) Int. J. Syst. Bacteriol. 38, 56–62. [Google Scholar]
- 6.Vandamme, P., Falsen, E., Rossau, R., Hoste, B., Segers, P., Tytgat, R. & De Ley, J. (1991) Int. J. Syst. Bacteriol. 41, 88–103. [DOI] [PubMed] [Google Scholar]
- 7.Wolin, M. J., Wolin, E. A. & Jacobs, N. J. (1961) J. Bacteriol. 81, 911–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simon, J., Gross, R., Klimmek, O. & Kröger A. (2000) in The Prokaryotes: An Evolving Electronic Resource for the Microbiological Community, eds. Balows, A., Trüper, H. G., Dworkin, M., Harder, W. & Schleifer, K. H. (Springer, New York), 3rd Ed.
- 9.Kröger, A., Biel, S., Simon, J., Gross, R., Unden, G. & Lancaster, C. R. D. (2002) Biochim. Biophys. Acta 1553, 23–38. [DOI] [PubMed] [Google Scholar]
- 10.Simon, J. (2002) FEMS Microbiol. Rev. 26, 285–309. [DOI] [PubMed] [Google Scholar]
- 11.Schumacher, W., Kroneck, P. M. H. & Pfennig, N. (1992) Arch. Microbiol. 158, 287–293. [Google Scholar]
- 12.Schuster, S. C. & Baeuerlein, E. (1992) J. Bacteriol. 174, 263–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Engelhardt, H., Schuster, S. C. & Baeuerlein, E. (1993) Science 262, 1046–1048. [DOI] [PubMed] [Google Scholar]
- 14.Schuster, S. C., Bauer, M., Kellermann, J., Lottspeich, F. & Baeuerlein, E. (1994) J. Bacteriol. 176, 5151–5155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schuster, S. C. & Khan, S. (1994) Annu. Rev. Biophys. Biomol. Struct. 23, 509–539. [DOI] [PubMed] [Google Scholar]
- 16.Fleischmann, R. D., Adams, M. D., White, O., Clayton, R. A., Kirkness, E. F., Kerlavage, A. R., Bult, C. J., Tomb, J. F., Dougherty, B. A., Merrick, J. M., et al. (1995) Science 269, 496–512. [DOI] [PubMed] [Google Scholar]
- 17.Gordon, D., Desmarais, C. & Green, P. (2001) Genome Res. 11, 614–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gordon, D., Abajian, C. & Green, P. (1998) Genome Res. 8, 195–202. [DOI] [PubMed] [Google Scholar]
- 19.Ewing, B., Hillier, L., Wendl, M. & Green, P. (1998) Genome Res. 8, 175–185. [DOI] [PubMed] [Google Scholar]
- 20.Ewing, B. & Green, P. (1998) Genome Res. 8, 186–194. [PubMed] [Google Scholar]
- 21.Meyer, F., Goesmann, A., McHardy, A. C., Bartels, D., Bekel, T., Clausen, J., Kalinowski, J., Linke, B., Rupp, O., Giegerich, R., et al. (2003) Nucleic Acids Res. 31, 2187–2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salzberg, S. L., Delcher, A. L., Kasif, S. & White, O. (1998) Nucleic Acids Res. 26, 544–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997) Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suzek, B. E., Ermolaeva, M. D., Schreiber, M. & Salzberg, S. L. (2001) Bioinformatics 17, 1123–1130. [DOI] [PubMed] [Google Scholar]
- 25.Tatusov, R. L., Koonin, E. V. & Lipman, D. J. (1997) Science 278, 631–637. [DOI] [PubMed] [Google Scholar]
- 26.Tomb, J.-F., White, O., Kerlavage, A. R., Clayton, R. A., Sutton, G. G., Fleischmann, R. D., Ketchum, K. A., Klenk, H. P., Gill, S., Dougherty, B. A., et al. (1997) Nature 388, 539–547. [DOI] [PubMed] [Google Scholar]
- 27.Alm, R. A., Ling, L.-S., Moir, D. T., King, B. L., Brown, E. D., Doig, P. C., Smith, D. R., Noonan, B., Guild, B. C., Dejonge, B. L., et al. (1999) Nature 397, 176–180. [DOI] [PubMed] [Google Scholar]
- 28.Parkhill, J., Wren, B. W., Mungall, K., Ketley, J. M., Churcher, C., Basham, D., Chillingworth, T., Davis, R. M., Feltwell, T., Holroyd, S., et al. (2000) Nature 403, 665–668. [DOI] [PubMed] [Google Scholar]
- 29.Pedersen, A. G., Jensen, L. J., Brunak, S., Stærfeldt, H. H. & Ussery, D. W. (2000) J. Mol. Biol. 299, 907–930. [DOI] [PubMed] [Google Scholar]
- 30.Lenger, R., Herrmann, U., Gross, R., Simon, J. & Kröger, A. (1997) Eur. J. Biochem. 246, 646–651. [DOI] [PubMed] [Google Scholar]
- 31.Simon, J. & Kröger, A. (1998) Arch. Microbiol. 170, 43–49. [DOI] [PubMed] [Google Scholar]
- 32.Bronder, M., Mell, H., Stupperich, E. & Kröger, A. (1982) Arch. Microbiol. 131, 216–223. [DOI] [PubMed] [Google Scholar]
- 33.Smith, M. A., Finel, M., Korolik, V. & Mendz, G. L. (2000) Arch. Microbiol. 174, 1–10. [DOI] [PubMed] [Google Scholar]
- 34.Lancaster, C. R. D. (2002) Biochim. Biophys. Acta 1553, 1–6. [DOI] [PubMed] [Google Scholar]
- 35.Ullmann, R., Gross, R., Simon, J., Unden, G. & Kröger, A. (2000) J. Bacteriol. 182, 5757–5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kelly, D. J. (2001) J. Appl. Microbiol. 90, 16S–24S. [DOI] [PubMed] [Google Scholar]
- 37.van Vliet, A. H. M., Ketley, J. M., Park, S. F. & Penn, C. W. (2002) FEMS Microbiol. Rev. 26, 173–186. [DOI] [PubMed] [Google Scholar]
- 38.Mahillon, J. & Chandler, M. (1998) Microbiol. Mol. Biol. Rev. 62, 725–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rakin, A., Schubert, S., Guilvot, I., Carniel, E. & Heesemann, J. (2000) FEMS Microbiol. Lett. 182, 225–229. [DOI] [PubMed] [Google Scholar]
- 40.Hacker, J. & Carniel, E. (2001) EMBO Rep. 2, 376–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koonin, E. V., Makarova, K. S. & Aravind, L. (2001) Annu. Rev. Microbiol. 55, 709–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jain, R., Rivera, M. C., Moore, J. E. & Lake, J. A. (2002) Theor. Popul. Biol. 61, 489–495. [DOI] [PubMed] [Google Scholar]
- 43.Bacon, D. J., Alm, R. A., Burr, D. H., Hu, L., Kopecko, D. J., Ewing, C. P., Trust, T. J. & Guerry, P. (2000) Infect. Immun. 68, 4384–4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Odenbreit, S., Püls, J., Sedlmaier, B., Gerland, E., Fischer, W. & Haas, R. (2000) Science 287, 1497–1500. [DOI] [PubMed] [Google Scholar]
- 45.Censini, S., Lange, C., Xiang, Z., Crabtree, J. E., Ghiara, P., Borodovsky, M., Rappuoli, R. & Covacci, A. (1996) Proc. Natl. Acad. Sci. USA 93, 14648–14653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Covacci, A., Telford, J. L., Del Giudice, G., Parsonnet, J. & Rappuoli, R. (1999) Science 284, 1328–1333. [DOI] [PubMed] [Google Scholar]
- 47.Hou, Y. M. (1999) Trends Biochem. Sci. 24, 295–298. [DOI] [PubMed] [Google Scholar]
- 48.Hacker, J. & Kaper, J. B. (2000) Annu. Rev. Microbiol. 54, 641–679. [DOI] [PubMed] [Google Scholar]
- 49.Ochman, H., Lawrence, J. G. & Groisman, E. A. (2000) Nature 405, 299–304. [DOI] [PubMed] [Google Scholar]
- 50.Konkel, M. E., Kim, B. J., Rivera-Amill, V. & Garvis, S. G. (1999) Mol. Microbiol. 32, 691–701. [DOI] [PubMed] [Google Scholar]
- 51.Wacker, M., Linton, D., Hitchen, P. G., Nita-Lazar, M., Haslam, S. M., North, S. J., Panico, M., Morris, H. R., Dell, A., Wren, B. W., et al. (2002) Science 298, 1790–1793. [DOI] [PubMed] [Google Scholar]
- 52.Szymanski, C. M., Yao, R., Ewing, C. P., Trust, T. J. & Guerry, P. (1999) Mol. Microbiol. 32, 1022–1030. [DOI] [PubMed] [Google Scholar]
- 53.Young, N. M., Brisson, J.-R., Kelly, J., Watson, D. C., Tessier, L., Lanthier, P. H., Jarrell, H. C., Cadotte, N., St. Michael, F., Aberg, E., et al. (2002) J. Biol. Chem. 277, 42530–42539. [DOI] [PubMed] [Google Scholar]
- 54.Hoch, J. A. & Silhavy, T. J., eds. (1995) Two-Component Signal Transduction (Am. Soc. Microbiol., Washington, DC).
- 55.Scoarughi, G. L., Cimmino, C. & Donini, P. (1999) J. Bacteriol. 181, 552–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kaneko, T., Nakamura, Y., Wolk, C. P., Kuritz, T., Sasamoto, S., Watanabe, A., Iriguchi, M., Ishikawa, A., Kawashima, K., Kimura, T., et al. (2001) DNA Res. 8, 227–253. [DOI] [PubMed] [Google Scholar]
- 57.Schultz, J., Milpetz, F., Bork, P. & Ponting, C. P. (1998) Proc. Natl. Acad. Sci. USA 95, 5857–5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ott, A., Oehme, F., Keller, H. & Schuster, S. C. (2000) EMBO J. 19, 5782–5792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shaulsky, G., Fuller, D. & Loomis, W. F. (1998) Development (Cambridge, U.K.) 125, 691–699. [DOI] [PubMed] [Google Scholar]
- 60.Tal, R., Wong, H. C., Calhoon, R., Gelfand, D., Fear A. L., Volman, G., Mayer, R., Ross, P., Amikam, D., Weinhouse, H., et al. (1998) J. Bacteriol. 180, 4416–4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.O'Toole, P. W., Lane, M. C. & Porwollik, S. (2000) Microbes Infect. 2, 1207–1214. [DOI] [PubMed] [Google Scholar]
- 62.Kennelly, P. J. (2002) FEMS Microbiol. Lett. 206, 1–8. [DOI] [PubMed] [Google Scholar]
- 63.Bergersen, F. J. & Hipsley, E. H. (1970) J. Gen. Microbiol. 60, 61–65. [DOI] [PubMed] [Google Scholar]
- 64.Rudnick, P., Meletzus, D., Green, A., He, L. & Kennedy, C. (1997) Soil Biol. Biochem. 29, 831–841. [Google Scholar]
- 65.Haselkorn, R., Lapidus, A., Kogan, Y., Vlcek, C., Paces, J., Paces, V., Ulbrich, P., Pecenkova, T., Rebrekov, D., Milgram, A., et al. (2001) Proc. Natl. Acad. Sci. USA 70, 43–52. [DOI] [PubMed] [Google Scholar]
- 66.Huang, T.-C., Lin, R.-F., Chu, M.-K. & Chen, H.-M. (1999) Microbiology 145, 743–753. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.