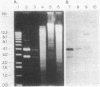
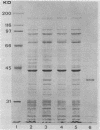
Abstract
Genes coding for three xylan-degrading activities, xylanase, xylosidase, and arabinosidase, were simultaneously cloned from the colonic anaerobic organism Bacteriodes ovatus. The genes for the three enzymes were located on a 3.8-kilobase EcoRI genomic insert and were cloned by using plasmid pUC18. All three activities were expressed in Escherichia coli JM83, and all were cell associated. Expression of the xylanase gene was independent from expression of the xylosidase and arabinosidase genes, whereas expression of the latter two genes appeared to be coordinated. Restriction endonuclease analysis of the arabinosidase and xylosidase genes and partial purification of these enzyme activities from E. coli suggested that these activities were catalyzed by a bifunctional protein or two proteins of very similar molecular weight. All three enzyme activities were regulated in B. ovatus in response to the carbon source used for growth. This is the first report of the cloning and expression of B. ovatus genes.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bigelis R., Keesey J. K., Fink G. R. The yeast his4 multifunctional protein. Immunochemistry of the wild type protein and altered forms. J Biol Chem. 1981 May 25;256(10):5144–5152. [PubMed] [Google Scholar]
- Bounias M. N-(1-naphthyl)ethylenediamine dihydrochloride as a new reagent for nanomole quantification of sugars on thin-layer plates by a mathematical calibration process. Anal Biochem. 1980 Aug;106(2):291–295. doi: 10.1016/0003-2697(80)90523-0. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Dev I. K., Harvey R. J. A complex of N5,N10-methylenetetrahydrofolate dehydrogenase and N5,N10-methenyltetrahydrofolate cyclohydrolase in Escherichia coli. Purification, subunit structure, and allosteric inhibition by N10-formyltetrahydrofolate. J Biol Chem. 1978 Jun 25;253(12):4245–4253. [PubMed] [Google Scholar]
- Greve L. C., Labavitch J. M., Hungate R. E. alpha-L-arabinofuranosidase from Ruminococcus albus 8: purification and possible role in hydrolysis of alfalfa cell wall. Appl Environ Microbiol. 1984 May;47(5):1135–1140. doi: 10.1128/aem.47.5.1135-1140.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III in DNA sequence analysis. Methods Enzymol. 1987;155:156–165. doi: 10.1016/0076-6879(87)55014-5. [DOI] [PubMed] [Google Scholar]
- Hespell R. B., Wolf R., Bothast R. J. Fermentation of xylans by Butyrivibrio fibrisolvens and other ruminal bacteria. Appl Environ Microbiol. 1987 Dec;53(12):2849–2853. doi: 10.1128/aem.53.12.2849-2853.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lee S. F., Forsberg C. W. Isolation and Some Properties of a beta-d-Xylosidase from Clostridium acetobutylicum ATCC 824. Appl Environ Microbiol. 1987 Apr;53(4):651–654. doi: 10.1128/aem.53.4.651-654.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panbangred W., Kawaguchi O., Tomita T., Shinmyo A., Okada H. Isolation of two beta-xylosidase genes of Bacillus pumilus and comparison of their gene products. Eur J Biochem. 1984 Jan 16;138(2):267–273. doi: 10.1111/j.1432-1033.1984.tb07911.x. [DOI] [PubMed] [Google Scholar]
- SAITO H., MIURA K. I. PREPARATION OF TRANSFORMING DEOXYRIBONUCLEIC ACID BY PHENOL TREATMENT. Biochim Biophys Acta. 1963 Aug 20;72:619–629. [PubMed] [Google Scholar]
- Salyers A. A., Gherardini F., O'Brien M. Utilization of xylan by two species of human colonic Bacteroides. Appl Environ Microbiol. 1981 Apr;41(4):1065–1068. doi: 10.1128/aem.41.4.1065-1068.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salyers A. A., Vercellotti J. R., West S. E., Wilkins T. D. Fermentation of mucin and plant polysaccharides by strains of Bacteroides from the human colon. Appl Environ Microbiol. 1977 Feb;33(2):319–322. doi: 10.1128/aem.33.2.319-322.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Weinstein L., Albersheim P. Structure of Plant Cell Walls: IX. Purification and Partial Characterization of a Wall-degrading Endo-Arabanase and an Arabinosidase from Bacillus subtilis. Plant Physiol. 1979 Mar;63(3):425–432. doi: 10.1104/pp.63.3.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Zanini V., Viviani M. A., Cava L., Tortorano A. M., Villa G. Candida infections in the burn patients. Panminerva Med. 1983 Jul-Sep;25(3):163–166. [PubMed] [Google Scholar]