Abstract
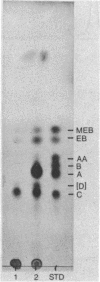
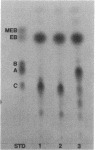
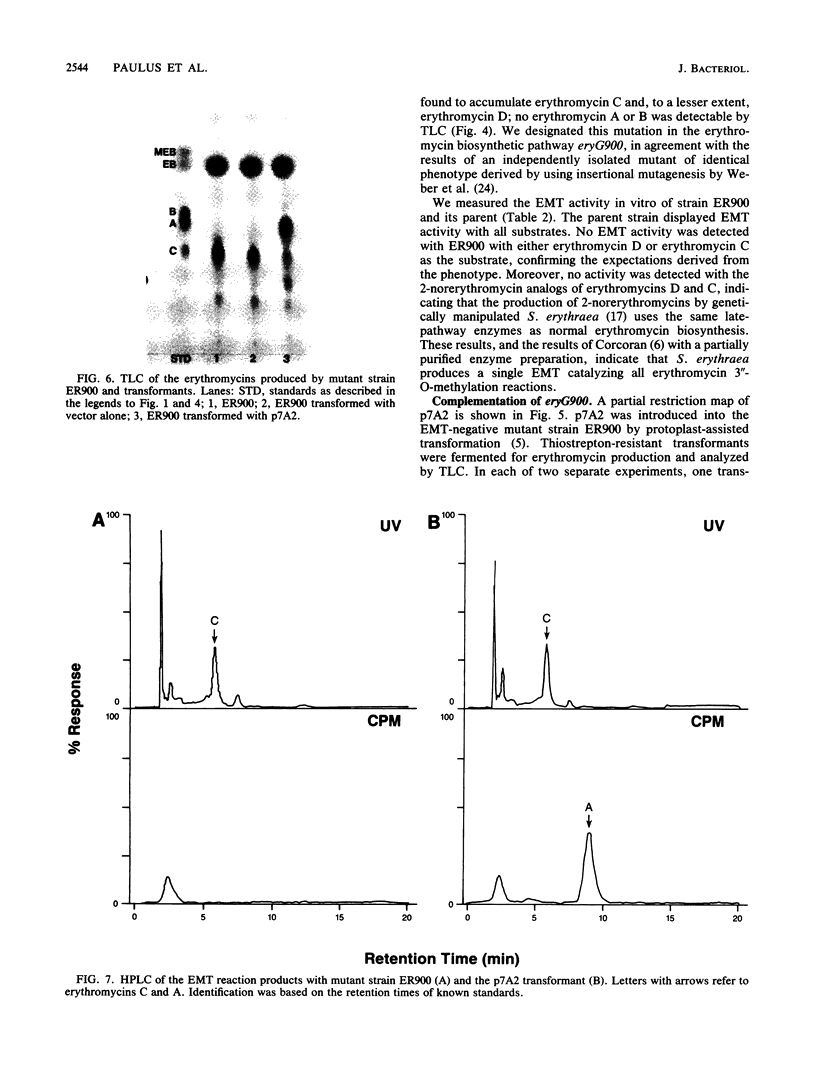
A mutant strain derived by chemical mutagenesis of Saccharopolyspora erythraea (formerly known as Streptomyces erythreus) was isolated that accumulated erythromycin C and, to a lesser extent, its precursor, erythromycin D, with little or no production of erythromycin A or erythromycin B (the 3"-O-methylation products of erythromycin C and D, respectively). This mutant lacked detectable erythromycin O-methyltransferase activity with erythromycin C, erythromycin D, or the analogs 2-norerythromycin C and 2-norerythromycin D as substrates. A 4.5-kilobase DNA fragment from S. erythraea originating approximately 5 kilobases from the erythromycin resistance gene ermE was identified that regenerated the parental phenotype and restored erythromycin O-methyltransferase activity when transformed into the erythromycin O-methyltransferase-negative mutant. Erythromycin O-methyltransferase activity was detected when the 4.5-kilobase fragment was fused to the lacZ promoter and introduced into Escherichia coli. The activity was dependent on the orientation of the DNA relative to lacZ. We have designated this genotype eryG in agreement with Weber et al. (J.M. Weber, B. Schoner, and R. Losick, Gene 75:235-241, 1989). It thus appears that a single enzyme catalyzes all of the 3"-O-methylation reactions of the erythromycin biosynthetic pathway in S. erythraea and that eryG codes for the structural gene of this enzyme.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baltz R. H., Seno E. T. Properties of Streptomyces fradiae mutants blocked in biosynthesis of the macrolide antibiotic tylosin. Antimicrob Agents Chemother. 1981 Aug;20(2):214–225. doi: 10.1128/aac.20.2.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basabe J. R., Lee C. A., Weiss R. L. Enzyme assays using permeabilized cells of Neurospora. Anal Biochem. 1979 Jan 15;92(2):356–360. doi: 10.1016/0003-2697(79)90670-5. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Brown D. P., Chiang S. J., Tuan J. S., Katz L. Site-specific integration in Saccharopolyspora erythraea and multisite integration in Streptomyces lividans of actinomycete plasmid pSE101. J Bacteriol. 1988 May;170(5):2287–2295. doi: 10.1128/jb.170.5.2287-2295.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran J. W. S-adenosylmethionine:erythromycin C O-methyltransferase. Methods Enzymol. 1975;43:487–498. doi: 10.1016/0076-6879(75)43109-3. [DOI] [PubMed] [Google Scholar]
- Hunaiti A. A., Kolattukudy P. E. Source of methylmalonyl-coenzyme A for erythromycin synthesis: methylmalonyl-coenzyme A mutase from Streptomyces erythreus. Antimicrob Agents Chemother. 1984 Feb;25(2):173–178. doi: 10.1128/aac.25.2.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majer J., Martin J. R., Egan R. S., Corcoran J. W. Antibiotic glycosides. 8. Erythromycin D, a new macrolide antibiotic. J Am Chem Soc. 1977 Mar 2;99(5):1620–1622. doi: 10.1021/ja00447a055. [DOI] [PubMed] [Google Scholar]
- Martin J. R., DeVault R. L., Sinclair A. C., Stanaszek R. S., Johnson P. A new naturally occurring erythromycin: erythromycin F. J Antibiot (Tokyo) 1982 Apr;35(4):426–430. doi: 10.7164/antibiotics.35.426. [DOI] [PubMed] [Google Scholar]
- Martin J. R., Perun T. J., Girolami R. L. Studies on the biosynthesis of the erythromycins. I. Isolation and structure of an intermediate glycoside, 3-alpha-L-mycarosylerythronolide B. Biochemistry. 1966 Sep;5(9):2852–2856. doi: 10.1021/bi00873a011. [DOI] [PubMed] [Google Scholar]
- Martin J. R., Rosenbrook W. Studies on the biosynthesis of the erythromycins. II. Isolation and structure of a biosynthetic intermediate, 6-deoxyerythronolide B. Biochemistry. 1967 Feb;6(2):435–440. doi: 10.1021/bi00854a010. [DOI] [PubMed] [Google Scholar]
- McAlpine J. B., Tuan J. S., Brown D. P., Grebner K. D., Whittern D. N., Buko A., Katz L. New antibiotics from genetically engineered actinomycetes. I. 2-Norerythromycins, isolation and structural determinations. J Antibiot (Tokyo) 1987 Aug;40(8):1115–1122. doi: 10.7164/antibiotics.40.1115. [DOI] [PubMed] [Google Scholar]
- Queener S. W., Sebek O. K., Vézina C. Mutants blocked in antibiotic synthesis. Annu Rev Microbiol. 1978;32:593–636. doi: 10.1146/annurev.mi.32.100178.003113. [DOI] [PubMed] [Google Scholar]
- Schulman M. D., Valentino D., Nallin M., Kaplan L. Avermectin B2 O-methyltransferase activity in "Streptomyces avermitilis" mutants that produce increased amounts of the avermectins. Antimicrob Agents Chemother. 1986 Apr;29(4):620–624. doi: 10.1128/aac.29.4.620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman M. D., Valentino D., Streicher S., Ruby C. "Streptomyces avermitilis" mutants defective in methylation of avermectins. Antimicrob Agents Chemother. 1987 May;31(5):744–747. doi: 10.1128/aac.31.5.744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seno E. T., Baltz R. H. S-Adenosyl-L-methionine: macrocin O-methyltransferase activities in a series of Streptomyces fradiae mutants that produce different levels of the macrolide antibiotic tylosin. Antimicrob Agents Chemother. 1982 May;21(5):758–763. doi: 10.1128/aac.21.5.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji K., Goetz J. F. HPLC as a rapid means of monitoring erythromycin and tetracycline fermentation processes. J Antibiot (Tokyo) 1978 Apr;31(4):302–308. doi: 10.7164/antibiotics.31.302. [DOI] [PubMed] [Google Scholar]
- Weber J. M., Schoner B., Losick R. Identification of a gene required for the terminal step in erythromycin A biosynthesis in Saccharopolyspora erythraea (Streptomyces erythreus). Gene. 1989 Feb 20;75(2):235–241. doi: 10.1016/0378-1119(89)90269-2. [DOI] [PubMed] [Google Scholar]
- Weber J. M., Wierman C. K., Hutchinson C. R. Genetic analysis of erythromycin production in Streptomyces erythreus. J Bacteriol. 1985 Oct;164(1):425–433. doi: 10.1128/jb.164.1.425-433.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]