Abstract
Disease resistance in transgenic plants has been improved, for the first time, by the insertion of a gene from a biocontrol fungus. The gene encoding a strongly antifungal endochitinase from the mycoparasitic fungus Trichoderma harzianum was transferred to tobacco and potato. High expression levels of the fungal gene were obtained in different plant tissues, which had no visible effect on plant growth and development. Substantial differences in endochitinase activity were detected among transformants. Selected transgenic lines were highly tolerant or completely resistant to the foliar pathogens Alternaria alternata, A. solani, Botrytis cinerea, and the soilborne pathogen Rhizoctonia solani. The high level and the broad spectrum of resistance obtained with a single chitinase gene from Trichoderma overcome the limited efficacy of transgenic expression in plants of chitinase genes from plants and bacteria. These results demonstrate a rich source of genes from biocontrol fungi that can be used to control diseases in plants.
Mycoparasitic and antagonistic fungi have been studied to develop a biological alternative or complement to chemical pesticides for the control of fungal pathogens (1). Strain selection and genetic mutation have produced some fungal isolates that are as effective as fungicides in certain culture conditions (2). The antifungal mechanism of Trichoderma spp., the best known biocontrol fungi, involves fungal cell wall degrading enzymes, such as chitinases and glucanases (3–7). The purified enzymes from T. harzianum, which have an antagonistic and a nutritional role, are strong inhibitors of many important plant pathogens and are able to lyse not only the “soft” structure of the hyphal tip but also the “hard” chitin wall of mature hyphae, conidia, chlamydospores, and sclerotia. They are substantially more antifungal than chitinolytic and glucanolytic enzymes purified thus far from any other source when assayed under the same conditions (i.e., up to 100 times more active than the corresponding plant enzymes and effective on a much wider range of pathogens) (3–7) and nontoxic to plants even at high concentrations. Furthermore, the antifungal activity is synergistically enhanced when different Trichoderma cell wall degrading enzymes are used together (5) or in combination with plant pathogenesis-related proteins (8), commercial fungicides, cell membrane-affecting toxins (3), or biocontrol bacteria (9). For these reasons, the genome of mycoparasites, which has evolved specifically to attack other fungi but not plants, represents a potential source of powerful antifungal genes.
The plant defense system against microbial pathogens may be modified to produce high constitutive levels of antimicrobial compounds (10). Plant genes encoding cell wall degrading enzymes, especially chitinases, have been used to alter plant resistance to fungal pathogens, but no single genes have produced an adequate level of resistance (11–14), and almost no papers report resistance to multiple pathogens. Reasons for this may be that plant chitinases: (i) usually affect only the hyphal tip and are unable to effectively degrade harder chitin structures, (ii) have weak antifungal activity alone, (iii) are inhibitory only to a limited number of fungal species, and (iv) have no effect on several important pathogens (15–17). In terms of antifungal activity, chitinase genes from biocontrol fungi such as Trichoderma are clearly an improvement over corresponding plant genes. These fungal genes encode for chitinolytic enzymes that can reach the antifungal activity level of some chemical fungicides based on ED50 values (3–5, 8, 9). Furthermore, extensive testing in vitro has shown that there are virtually no chitinous pathogens resistant to Trichoderma chitinases (refs. 3–5, 8, and 9 and M.L., G.E.A., and S.L.W., unpublished work). Therefore, it is expected that the transgenic use of these enzymes should produce a high level of resistance in crop plants against a variety of fungal pathogens and, in contrast to plant genes, could be accomplished with a single fungal gene.
Recently, we cloned from T. harzianum the gene ThEn-42 (chit42) (18, 19) that encodes a powerful endochitinase with a much stronger antifungal activity vs. a wide range of phytopathogens compared with other chitinolytic enzymes (3–6, 8). Constitutive expression of the T. harzianum gene was obtained in tobacco and potato, and transgenic lines showed a high level and a broad range of resistance against both soil-borne and foliar pathogens. Strategies based on the use of “mycoparasitic genes” to improve genetically plant disease resistance are discussed.
MATERIALS AND METHODS
Constructs and Plant Transformation.
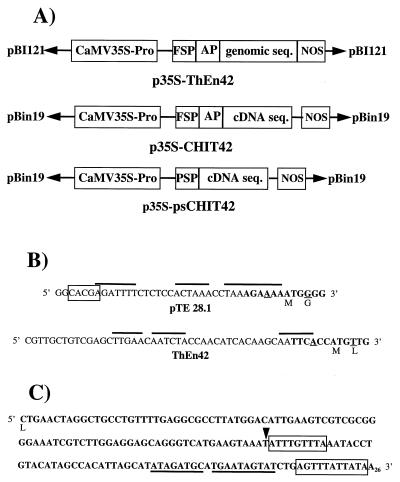
The endochitinase-encoding genes ThEn-42 and chit42 were cloned from T. harzianum strain P1 (ATCC 74058) and strain CECT (Coleccion Española de Cultivos Tipo) 2413, respectively (18, 19). Both genes encode for the same antifungal endochitinase (3) and have >90% DNA sequence homology. Binary vectors were constructed that contained ThEn-42 and chit42 under the control of the cauliflower mosaic virus 35S subunit (CaMV35S) promoter region and the Agrobacterium nopaline synthase terminator (Fig. 1A). A genomic copy of ThEn-42, which includes a 26-aa secretion peptide followed by a 12-aa putative activation peptide and a 424-aa coding sequence interrupted by three introns (19), was ligated into pBI121 to produce the plasmid p35S-ThEn42 (Fig. 1A). To express constitutively the chit42 cDNA, two constructs were made in the plasmid pBin19:p35S-CHIT42 containing the Trichoderma signal and putative activation peptides and p35S-psCHIT42 containing a tomato pathogenesis-related protein (P1-p14) signal peptide from pTE 28.1 (20) (Fig. 1 A and B). In the latter, the 3′ noncoding region of the fungal gene was modified to improve transcription and mRNA stability (Fig. 1C) by removing a terminal sequence of ≈100 bp. The transformation procedure followed standard protocols (21): Agrobacterium tumefaciens strain LBA4404 containing vectors with the chimeric fungal endochitinase gene and corresponding empty vectors was used to transform leaf disks of Nicotiana tabacum cv. Samsun NN and cv. Xhantii and stem segments of Solanum tuberosum cv. Desiree (only p35S-ThEn42).
Figure 1.
Cassettes containing ThEn42 (genomic or cDNA) used for plant transformation. (A) CaMV35S-Pro = promoter; nopaline synthase (NOS) = transcription terminator; coding sequence for the fungal signal peptide (FSP), plant secretion peptide (PSP), fungal putative activation peptide (AP). (B) The 5′ noncoding region of p35S-psCHIT42 with PSP (pTE 28.1) and p35S-CHIT42 with FSP. Box indicates mRNA stabilization consensus sequence, and topline indicates A+T rich regions (20). Bold letters indicate consensus sequences for transcription in the ATG region compared with TAA ACA ATG GCT (33). (C) Elements reducing mRNA stability at the 3′ noncoding region of the fungal gene (33) are boxed or underlined. In p35S-psCHIT42, the region downstream from restriction site SspI (arrowhead) was removed.
Screening and Molecular Analyses of Transgenic Lines.
Kanamycin-resistant transgenic plants were propagated on Murashige–Skoog basal salt medium (Sigma) containing sucrose (3%) and kanamycin sulfate (100 mg/liter). Homozygotic lines were selected by selfing F1 progeny. Plant and fungal DNA were obtained following published procedures (22, 23). The transgene in selected progeny was PCR-amplified with the primers GGTTATGCTTTCCATCGG (EC1), GTGGTAGCCCGGAGCGTATT (Bio1N) and CAAGGAGTCAGAGCCAGTCTT(BB2), which anneal at 566 bp (EC1), 813 bp (Bio1N), and 1,367 bp (BB2) from the 5′ end of the gene (Fig. 2A). The copy number of the endochitinase gene was assessed by Southern analysis (24) of transformant DNA cleaved with BamHI or EcoRI, followed by a test of segregation in the F1 generation. The DNA fragments used as probes were obtained from EcoRI digestion of p35S-ThEn42 and p35S-CHIT42 and were labeled (19, 25). For Northern analysis, 10 μg of RNA were used following published methods (26, 27). ELISA tests and Western analysis with antibodies prepared against the purified endochitinase from T. harzianum (18, 28) were performed on protein extracts from leaves, stems, flowers, and roots of selected transgenic lines (29). The transgenic protein also was detected and quantified by using turbidity reduction and fluorescent assays with specific substrates (4-methylumbelliferyl-β-d-N,N′,N"-triacetylchitotrioside and nitrophenyl-β-d-N,N′,N"-triacetylchitotrioside) as described (30, 31). Localization of the transgenic protein in the plant tissues was performed by immunogold labeling following standard fixation and by electron microscopy observation.
Figure 2.
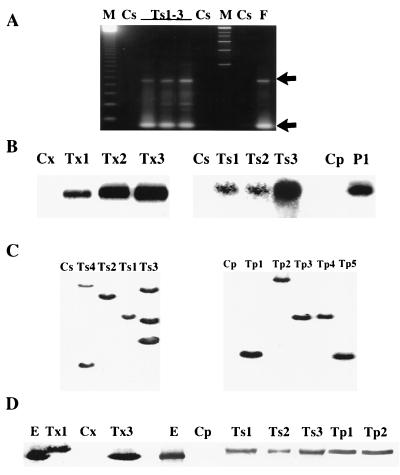
Analyses of representative transgenic (T) tobacco and potato lines. C, control plants (nontransformed or empty-vector transformed): Cs, tobacco cv. Samsun NN; Cx, tobacco cv. Xhantii; Cp, potato cv. Desiree. Representative samples of transgenic lines are shown: Ts(no. of plant line) = tobacco cv. Samsun NN transformed with p35S-ThEn42; Tx (no. of plant line) = tobacco cv. Xhantii transformed with p35S-CHIT42 (Tx1) or with p35S-psCHIT42 (Tx2 and Tx3); Tp(no. of plant line) = potato cv. Desiree transformed with p35S-ThEn42; E = pure endochitinase from T. harzianum culture. (A) PCR amplification of the fungal transgene using transgenic, control plant DNA or control fungal DNA (F) with nested primer sets EC1-Bio1N and EC1-BB2 producing 250- and 800-bp bands, respectively (indicated by arrows) (18). (B) Northern analysis of plant tissues using the ThEn-42 cDNA sequence as a probe and visualization of bands after 3–5 days of exposure. (C) Southern analysis of BamHI-digested tobacco and potato DNA probed with an EcoRI 1.7-kb fragment of ThEn-42. (D) Western analysis of soluble leaf proteins of tobacco and potato separated by SDS/PAGE and visualized with anti-endochitinase antibodies and a standard alkaline phosphatase immunoassay.
Disease Resistance Assays.
Transgenic tobacco and potato plants were tested for resistance to: Alternaria alternata isolate FC2600 and A. solani isolate 1191 from potato, airborne foliar pathogens that cause brown spot or leaf spot disease; Botrytis cinerea isolate 319 from tobacco, an airborne pathogen that causes grey mold disease; and Rhizoctonia solani from tomato (isolate 1556), an endemic soilborne pathogen that causes damping-off, seedling blight, and root rot. All isolates were from the University of Naples culture collection.
Assays with A. alternata, A. solani, and B. cinerea.
Transgenic and control tobacco were grown at 25°/15°C with 16 h of diurnal light in the greenhouse. Four leaves per plant from 10 plants/line of both cultivars were inoculated with 10 μl of a spore suspension (106/ml), obtained from A. alternata grown on potato dextrose agar (25°C with light), at four inoculation points between the leaf veins; and the plants were incubated at 20°C with 16 h of diurnal light and 90% relative humidity (RH). Observations were conducted daily (up to 11 days) for disease reaction (chlorosis, necrosis, or both) and severity (none, slight, strong, severe).
Ten potato plants/line were spray inoculated with A. solani conidia (104/ml) obtained on potato dextrose agar (32), incubated at 100% RH for 16 h, and allowed to dry, and the procedure was repeated three times. Ten days after inoculation, the number of lesions/leaf was evaluated per plants/line and averaged.
Four leaves per plant from 10 plants/line of both tobacco cultivars were inoculated with B. cinerea agar plugs (0.5–1 cm2) from 1-week-old cultures grown on malt extract agar, and plants were incubated at 18°C with 100% RH. Lesion sizes were measured 10 and 14 days after inoculation, and results were combined and averaged for each plant line.
Assays with R. solani.
For all R. solani assays, the fungus was grown on potato dextrose broth at 150 rpm, 25°C with light for 5 days, and the mycelium was vacuum harvested, weighed, and homogenized in distilled water. In one set of seedling assays, a water agar solution containing the fungal suspension (0.7% agar, 0–1.0 mg fungus/ml) was prepared in Petri plates. One-month-old tobacco seedlings (25 plants/line), germinated on Murashige–Skoog with 0.7% agar, were transferred to the fungus-containing or noncontaining plates. In the other seedlings assays, sterile soil (1 cm deep) was placed in containers (115 mm diameter × 35 mm deep) and infested by evenly applying a 10-ml R. solani suspension (0.75 mg/ml) to the surface and then covered with a layer of noninoculated soil (1.5 cm deep). Seedlings grown to emergence of the second set of true leaves (20 plants/line) were transplanted into the surface layer, misted, and covered with a transparent lid. For both assays, plants were incubated at 20°C with 16 h of diurnal light at 90% RH. The number of dead or dying plants was counted every 3 days.
For assays with grown plants, tobacco (10 plants/line) was grown in peat moss pellets until the 5–6 leaf stage (2 months old) and then transplanted into boxes (20 × 20 × 5 cm3 or 35 × 26 × 9 cm3) containing 1 or 3 liters of sterile soil, respectively. A R. solani suspension (1–3 g/liter) was prepared and watered around the base of the plants, followed by incubation at 20°C with 16 h of diurnal light at 90% RH. Evaluations were conducted weekly, up to 1.5 months after transplant, for disease symptoms and effect on plant growth.
Potato plants were micropropagated on Murashige–Skoog media plus 0.3% sucrose with 0.7% agar, until root formation, then individually transplanted to 50-ml tubes containing sterile soil and climatized for 2 weeks. Each plant (10 plants/line) was inoculated with a 10-ml R. solani suspension (5 mg/ml) and then grown at 20°C with 16 h of diurnal light at 90% RH. Evaluations were conducted twice weekly for overall plant condition and measurement of plant height.
For all disease resistance assays, infected plant material was plated onto acidified potato dextrose agar to verify the presence of the test fungus. To maintain virulence, all pathogens were continuously re-isolated from infected plants. Each experiment contained several replicates and was repeated three or more times. Presented results consisted of combined data for each tobacco or potato line, with calculations of means and SDs (indicated with bars in the figures). Data also were combined for all lines and analyzed by ANOVA.
RESULTS
Transgenic Expression of the Trichoderma Chitinase Gene in Tobacco and Potato.
Fifty tobacco and 13 potato kanamycin-resistant transformants were screened for the presence and expression of the fungal endochitinase gene by PCR, Northern, Southern, and Western analyses (Fig. 2 A–D). Most transformed plants but not nontransformed and empty vector-transformed plants contained the transgene, the mRNA of the expected size, and a protein that reacted with antibodies raised against the Trichoderma endochitinase (18, 28) (Fig. 2 A–D). Southern analysis indicated that the copy number of the transgene was one to three and occasionally more, and there was no correlation between the number of copies and the level of transcription in transgenic lines (data not shown).
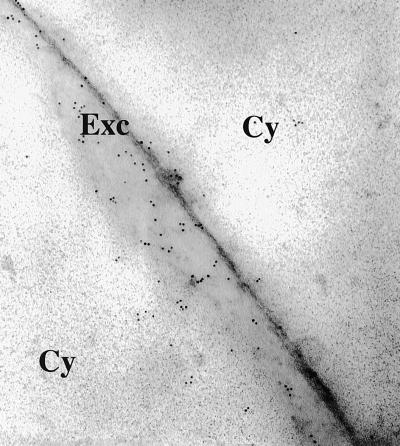
The transgenic protein accumulated mainly in leaves and stems, but also was found in roots and flowers. The protein encoded by the transgene was identical to the wild-type Trichoderma endochitinase in plants transformed with p35S-psCHIT42 but was slightly larger in plants transformed with p35S-ThEn42 and p35S-CHIT42 (Fig. 2D) because the 12-aa putative activation peptide at the 5′ end of the gene (19) was not cleaved. However, both the Trichoderma and tomato secretion signal peptides were cleaved and induced extracellular accumulation of the fungal enzyme as shown by immunogold labeling (Fig. 3). Specific enzyme assays and protein band image analysis indicated that the fungal endochitinase was active in both transgenic tobacco and potato, and, depending on the lines tested, it represented 0.01–0.5% of total protein content. Specific enzyme activity, pH, and temperature optima and stability of the transgenic enzyme showed no significant changes in comparison to the purified Trichoderma endochitinase (data not shown).
Figure 3.
Gold immunolabeling of transgenic tobacco cv. Samsun NN (Ts1) transformed with p35S-ThEn42. Cy, cytoplasm; Exc, extracellular space.
In general, the highest endochitinase activity was found in tobacco and potato bearing the genomic copy (p35S-ThEn42) and in tobacco with the cDNA fused to the tomato secretion sequence (p35S-psCHIT42). Progenies reached 10- and 400-fold more endochitinase activity than control plants in roots and leaves, respectively, although intermediate, low, or nondetectable levels also were found. Tobacco and potato plants with high chitinase activity showed no obvious differences in germination, growth, or development when compared with the controls or wild types. In addition, up to 1 mg of purified endochitinase from T. harzianum administered by intervenal injection had no effect on tobacco and potato leaves (data not shown).
Disease Resistance of Transgenic Plants to Foliar Pathogens.
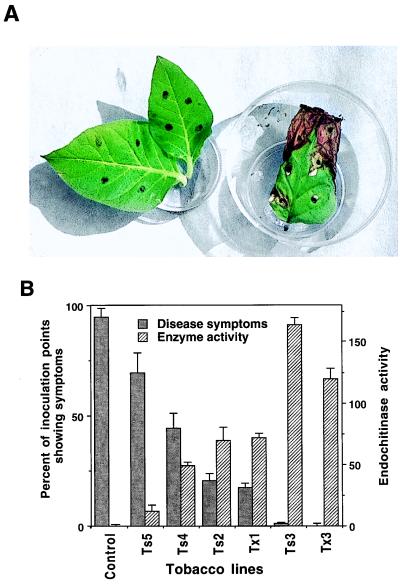
Thirty-eight transgenic lines of tobacco and nine of potato were assayed. Tobacco controls showed typical symptoms of A. alternata infection at 96% of the inoculation sites (Table 1) whereas the overall value for all transgenics was 34%. Controls showed typical necrosis surrounded by a chlorotic halo and extensive sporulation of the pathogen whereas symptoms on transgenics, if present, were a faint chlorosis or a hypersensitive-like necrosis with little sporulation (data not shown). The level of disease resistance varied among transgenic lines, from no to complete resistance, but the average reaction to inoculation ranged from no (66%) to slight reaction (34%) (Table 1). Only line Ts5 exhibited a strong or severe reaction like the controls, and it was not considered in the overall evaluation of transformants (Table 1). Approximately 5–10% of the transgenic lines were completely resistant to A. alternata (Fig. 4 A and B; Table 1; and data not shown) and never developed symptoms even if exposed to the pathogen repeatedly and for long periods. Generally, transgenic plants expressing higher amounts of Trichoderma endochitinase were more tolerant or completely resistant to A. alternata (Fig. 4B).
Table 1.
Resistance of Trichoderma endochitinase tobacco plants to A. alternata
| Plant lines | Disease symptoms
|
Disease severity
|
|||||
|---|---|---|---|---|---|---|---|
| R | C | N | None | Slight | Strong | Severe | |
| Ts2 | 20.5 | 18.1 | 15.7 | 79.5 | 20.5 | 0.0 | 0.0 |
| Ts3 | 1.0 | 0.0 | 1.0 | 99.0 | 1.0 | 0.0 | 0.0 |
| Ts4 | 44.3 | 15.7 | 37.1 | 55.7 | 44.3 | 0.0 | 0.0 |
| Ts5 | 69.5 | 64.2 | 66.8 | 30.5 | 28.5 | 22.5 | 18.5 |
| Tx1 | 17.5 | 17.5 | 0.0 | 82.5 | 17.5 | 0.0 | 0.0 |
| Tx3 | 0.0 | 0.0 | 0.0 | 100.0 | 0.0 | 0.0 | 0.0 |
| All trans. | 33.6 | 29.3 | 25.9 | 66.4 | 33.6 | 0.0 | 0.0 |
| Cs1 | 93.5 | 90.3 | 83.9 | 6.5 | 48.3 | 45.2 | 0.0 |
| Cs2 | 100.0 | 100.0 | 83.3 | 0.0 | 0.0 | 0.0 | 100.0 |
| Cs3 | 95.7 | 95.7 | 95.7 | 4.3 | 32.6 | 45.7 | 17.4 |
| Cx1 | 100.0 | 100.0 | 66.7 | 0.0 | 0.0 | 0.0 | 100.0 |
| Cx2 | 100.0 | 100.0 | 100.0 | 0.0 | 0.0 | 0.0 | 100.0 |
| All contr. | 95.8 | 94.7 | 89.5 | 4.2 | 31.6 | 36.8 | 27.4 |
Disease symptoms and severity (5 days after inoculation) of representative transgenic and control lines (see legend to Fig. 2) are indicated as percentage of inoculation points.
All trans., combined values of all transgenic lines (except line Ts5) tested; All contr., combined values of all control lines tested; R, any reaction (C, N, or C + N); C, chlorosis; N, necrosis.
Figure 4.
Resistance of Trichoderma endochitinase tobacco plants to A. alternata. (A) Leaves of transgenic (line Ts3, on left) and control (on right) tobacco; dark dots indicate inoculation points. (B) Disease symptoms 11 days after inoculation and endochitinase activity in leaf protein extracts of representative transgenic lines (see legend to Fig. 2); control is an average of different lines of Cs and Cx; endochitinase activity was expressed as pmol of 4-methylumbelliferone released from 4-methylumbelliferyl-β-d-N-N′-N"-triacetylchitotriose/min/μg of protein (30–31).
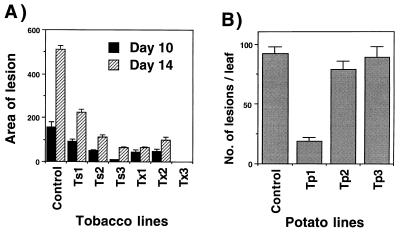
Assays with B. cinerea indicated that transgenic tobacco were highly tolerant and, in a few cases, nearly completely resistant to this pathogen. Generally, lesions were much larger in control plants than in transgenic lines expressing high levels of endochitinase activity. Selected lines, such as lines Tx3 and Ts3, that showed high levels of resistance to A. alternata, were also highly resistant to B. cinerea (Figs. 4B and 5A).
Figure 5.
Resistance of Trichoderma endochitinase tobacco plants to B. cinerea and of potato plants to A. solani. (A) Size of lesions (mm2) produced on leaves of different tobacco transgenic lines (see legend to Fig. 2) and controls (average of Cs and Cx lines) 10 and 14 days after inoculation with B. cinerea agar plugs. (B) Number of lesions observed on different potato transgenic lines and control (see legend to Fig. 2) 9 days after spray inoculation with A. solani.
Potato control plants showed typical disease symptoms when infected by A. solani, whereas the level of resistance varied greatly among transgenic lines (Fig. 5B). One-third of the potato lines were highly resistant to A. solani (i.e., line Tp1 showed 85% disease reduction), even under assay conditions highly favorable to disease development (Fig. 5B). As in tobacco, potato lines producing more Trichoderma endochitinase (about 200-fold more than controls) were more resistant to Alternaria (data not shown).
Disease Resistance of Transgenic Plants Against a Soil-Borne Pathogen.
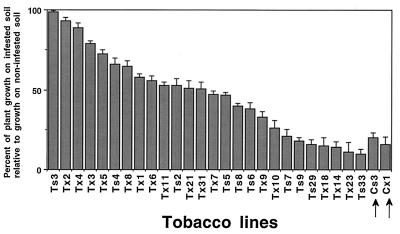
In experiments on water agar containing tobacco seedlings and R. solani, the pathogen was highly virulent and killed most of the control plants (Fig. 6A). Instead, survival rates among transgenics ranged from 65 to 80%. In soil experiments with Rhizoctonia and tobacco seedlings, 10–15% of the transgenic lines consistently showed no disease symptoms whereas others had little or no improvement in disease resistance (data not shown). A strong difference between controls and transgenic tobacco also was observed when 2-month-old plants were tested (Figs. 6B and 7). Growth of controls in R. solani-infested soil was reduced ≈90% in comparison to growth on noninfested soil. Several transgenic lines consistently performed better than controls and grew equally well as plants on noninfested soil, showing complete resistance (Fig. 7). Analysis of all of the transgenic lines combined indicated a significant increase in plant growth in comparison to the controls (P < 0.05) (data not shown). Surviving control plants grown in infested soil were noticeably stunted, whereas the Trichoderma endochitinase plants usually appeared larger and with more root regrowth (data not shown).
Figure 6.
Resistance of Trichoderma endochitinase tobacco plants to R. solani. (A) Seedlings grown 5 days on water agar containing R. solani: controls (Cs) without pathogen (1) or with pathogen (2); transgenics (Ts3) without pathogen (3) or with pathogen (4). (B) Transgenic and control plants grown on soil infested with R. solani; best standing plants are transgenic (boxes on right and asterisks) compared with plants grown on noninfected soil (left corner).
Figure 7.
Resistance of Trichoderma endochitinase tobacco plants to R. solani. Plant growth from 14 to 30 days on R. solani-infested soil relative to growth on noninfested soil (expressed as a percentage of plant height obtained on noninfested soil, which was taken as 100%). Control plants (indicated by arrows) and representative transgenic lines (see legend to Fig. 2); Tx (2 to 10 and 14) transformed with p35S-psCHIT42; Tx (1, 11, 18, 21, 23, and 31) transformed with p35S-CHIT42.
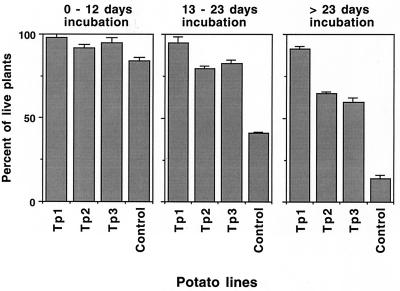
In disease tests of potato with R. solani, almost all of the control plants were killed after 23 days of incubation, and the few surviving plants appeared damaged and unhealthy. Most of the transgenic lines, five of nine tested, were considerably more resistant than controls, although a variation in disease resistance was observed (Fig. 8). A few potato lines, including Tp1, which was highly resistant to A. solani, consistently showed a complete or a nearly complete resistance to R. solani (Fig. 8).
Figure 8.
Resistance of Trichoderma endochitinase potato plants to R. solani. Percent survival of control and representative transgenic lines (see legend to Fig. 2) grown on R. solani-infested soil measured at different incubation periods.
DISCUSSION
The endochitinase gene from T. harzianum was expressed in tobacco and potato at high levels, and both secretion peptides from fungus and tomato were correctly cleaved and able to drive the accumulation of the transgenic enzyme outside the plant cell, which indicated that genes and signal sequences from fungi may be used to secrete and accumulate fungal enzymes in plants or in media containing plant cell suspensions. The Trichoderma introns of the genomic copy (p35S-ThEn42) were spliced correctly in plant. The level of transcription obtained with the genomic copy was higher than with the original cDNA copy and as high as with the cDNA engineered to improve mRNA stability. This result suggests that the presence of introns and the absence of other noncoding fungal sequences may improve gene expression in plants (33, 34).
Many papers and reviews have discussed the advantages of using chitinases for plant protection because these enzymes are fungicidal, part of the plant defense system, and nontoxic to plants, animals, and higher vertebrates. Transgenic expression in plant of chitinase genes has been shown to improve disease resistance in various crops, and patents have been issued on related methods. However, the level and the spectrum of resistance obtained has not supported the development of this technology for the production of new disease-resistant varieties suitable for commercial agriculture. One of the main limitations has been the relatively low level of resistance obtained with a single chitinase gene, which has resulted in the need to use gene combinations. This strategy, however, may not be attractive because a higher degree of genetic modification of the plant may substantially increase the costs of product development, complicate and delay the registration process in some countries, and be in contrast with the public’s feelings about accepting genetically modified crops. In addition, there are almost no reports of chitinase-transgenic plants showing resistance to several fungi, indicating that the genes and enzymes used so far have a narrow spectrum of antifungal activity. This spectrum may represent a major limitation for protecting crops that are susceptible to more than one pathogen and makes this transformation-based strategy less suitable to substitute or integrate the application of chemical fungicides that have a relatively broad spectrum of activity. The unsuccessful exploitation of chitinases for plant protection probably comes from the fact that the genes used so far, mostly from plants, code for enzymes that lack a strong inhibitory activity at low concentrations and the ability to efficiently degrade spores or other fungal preservation structures that serve as inoculum. The effectiveness of plant chitinases depends on the simultaneous action of other antifungal substances, and plant pathogens that have been exposed to these enzymes have developed mechanisms to overcome the defensive action of these chitinases (35). In fact, Cladosporium fulvum, Cercospora nicotiana, C. beticola, Fusarium solani, and several other pathogenic fungi were not inhibited in the growth by treatment with chitinases obtained from plant (15, 17).
This work demonstrates a successful application of a single endochitinase-encoding gene from T. harzianum for controlling plant diseases. Expression of ThEn-42 in tobacco and potato produced complete or nearly complete resistance to R. solani, A. solani, B. cinerea, and A. alternata, which was genetically stable and transferred to the progeny. This high level of resistance to both foliar and soilborne pathogens represents a major improvement in comparison to results from transgenic expression of other chitinase genes, i.e., from plants or bacteria. This improvement is in accordance with the finding that chitinases and other cell wall degrading enzymes from Trichoderma have a stronger and wider spectrum of antifungal activity in comparison to corresponding plant and bacterial enzymes (4). Conceivably, a number of useful genes may be found in these antagonistic fungi that developed a multi-component biochemical arsenal to survive in the highly competitive environment of the phyllosphere and rhizosphere. Therefore, this work discovers a new source of genes that can be used either singly or in synergistic combinations to provide protection against a wide range of fungal pathogens.
The level of disease resistance observed varied greatly among transgenic lines, but ≈5–10% of the transformed tobacco and potato plants were highly resistant to all the pathogens tested. These results obtained with one transgene may be explained by the following points: (i) there was a very high level of chitinolytic activity in some transgenic lines; (ii) the endochitinase concentrated in the extracellular space had a strong antifungal activity; (iii) the Trichoderma enzyme is highly synergistic with antifungal pathogenesis-related proteins in plants, such as tobacco osmotin (8); and (iv) the Trichoderma endochitinase in plant tissues may release from the cell wall of invading fungi compounds that elicit the plant defense response. Ongoing experiments support this latter hypothesis because potent elicitors that produce resistance responses in wild-type tobacco can be obtained by treating the mycelium of R. solani and Alternaria spp. with purified Trichoderma chitinolytic enzymes (M.L. and S.L.W., unpublished work). Moreover, in some cases, A. alternata inoculated on transgenic leaves produced a hypersensitive-like reaction. However, the level of resistance to the three foliar pathogens corresponded to the level of chitinolytic activity of the plant, indicating that the antifungal/lytic activity of the Trichoderma enzymes had a major role in the observed resistance.
A full scale application of fungal biocontrol agents in commercial agriculture has been delayed because of the inconsistent results obtained by introducing these complex microorganisms into the ever-changing environment. We suggest a way to overcome the often troublesome and unpredictable application of the whole organism by transferring directly into the plant the microbial genes responsible for disease control.
Acknowledgments
We are grateful to Kristen Ondik, Paolo Gori, Silvana Esposito, Rosa Verde, and Vincenzo Manzo for their help and assistance. S.L.W. has been supported by grants from Consorzio per la Ricerca Applicata in Agricoltura (CRAA) and Piano Nazionale Biotecnologie Vegetali del Mi.R.A.A.F., and M.L. has been supported in part by a Fullbright fellowship for research.
ABBREVIATION
- RH
relative humidity
References
- 1.Weindling R. Phytopathology. 1934;24:1153–1179. [Google Scholar]
- 2.Harman G E, Hayes C K. Report to the Office of Technology Assessment. Washington, D.C.: U.S. Congress; 1996. pp. 34–96. [Google Scholar]
- 3.Lorito M, Peterbauer C, Hayes C K, Harman G E. Microbiology. 1994;140:623–629. doi: 10.1099/00221287-140-3-623. [DOI] [PubMed] [Google Scholar]
- 4.Lorito M, Harman G E, Hayes C K, Broadway R M, Tronsmo A, Woo S L, Di Pietro A. Phytopathology. 1993;83:302–307. [Google Scholar]
- 5.Lorito M, Hayes C K, Di Pietro A, Woo S L, Harman G E. Phytopathology. 1994;84:398–405. [Google Scholar]
- 6.Lorito M, Farkas V, Rebuffat S, Bodo B, Kubicek C P. J Bacteriol. 1996;178:6382–6385. doi: 10.1128/jb.178.21.6382-6385.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lorito M, Peterbauer C, Sposato P, Mach R L, Strauss J, Kubicek C P. Proc Natl Acad Sci USA. 1996;93:14868–14872. doi: 10.1073/pnas.93.25.14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lorito M, Woo S L, D’Ambrosio M, Harman G E, Hayes C K, Kubicek C P, Scala F. Mol Plant-Microbe Interact. 1996;9:206–213. [Google Scholar]
- 9.Lorito M, Di Pietro A, Hayes C K, Woo S L, Harman G E. Phytopathology. 1993;83:721–728. [Google Scholar]
- 10.Staskawicz B J, Ausubel F M, Baker B J, Ellis J G, Jones J D G. Science. 1995;268:661–667. doi: 10.1126/science.7732374. [DOI] [PubMed] [Google Scholar]
- 11.Broglie K, Chet I, Holliday M, Cressman R, Biddle P, Knowlton S, Mauvais C J, Broglie R. Science. 1991;254:1194–1197. doi: 10.1126/science.254.5035.1194. [DOI] [PubMed] [Google Scholar]
- 12.Punja Z K, Raharjo S H T. Plant Dis. 1996;80:999–105. [Google Scholar]
- 13.Zhu Q, Maher E A, Masoud A, Dixon R A, Lamb C J. Bio/Technology. 1994;12:807–812. [Google Scholar]
- 14.Jach G, Görnhardt B, Mundy J, Logemann J, Pinsdorf E, Leach R, Schell J, Maas C. Plant J. 1995;8:97–109. doi: 10.1046/j.1365-313x.1995.08010097.x. [DOI] [PubMed] [Google Scholar]
- 15.Joosten M H A J, Verbakel H M, Nettekoven M E, van Leeuwen J, van der Vossen R T M, de Wit P J G M. Physiol Mol Plant Pathol. 1995;46:45–49. [Google Scholar]
- 16.Neuhaus J-M, Ahl-Goy P, Hinz U, Flores S, Meins F. Plant Mol Biol. 1991;16:141–151. doi: 10.1007/BF00017924. [DOI] [PubMed] [Google Scholar]
- 17.Mauch F, Mauch-Mani B, Boller T. Plant Physiol. 1988;88:936–942. doi: 10.1104/pp.88.3.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hayes C K, Klemsdal S, Lorito M, Di Pietro A, Peterbauer C, Nakas J P, Tronsmo A, Harman G E. Gene. 1994;138:143–148. doi: 10.1016/0378-1119(94)90797-8. [DOI] [PubMed] [Google Scholar]
- 19.Garcia I, Lora J M, de la Cruz J, Benitez T, Llobell A, Pintor-Toro J A. Curr Genet. 1994;27:83–89. doi: 10.1007/BF00326583. [DOI] [PubMed] [Google Scholar]
- 20.Vera P, Conejero V. J Gen Virol. 1989;70:1933–1942. [Google Scholar]
- 21.Horsh R B, Fry J E, Hoffman N C, Eichhltz D, Rogers S G, Fraley R T. Science. 1985;227:1229–1231. [Google Scholar]
- 22.Doyle J J, Doyle J L. Focus. 1994;12:1–10. [Google Scholar]
- 23.Raeder U, Broda P. Lett Appl Microbiol. 1985;1:17–20. [Google Scholar]
- 24.Church G M, Gilbert W. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning, A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 26.Pawlowski K, Kunze R, De Vries S, Bisseling T. Plant Mol Biol Manual. 1994;D 5:1–13. [Google Scholar]
- 27.Wadsworth G H, Redinbaugh M H, Scandalios J G. Anal Biochem. 1988;172:279–283. doi: 10.1016/0003-2697(88)90443-5. [DOI] [PubMed] [Google Scholar]
- 28.De la Cruz J, Hidalgo-Gallego A, Lora J M, Benítez T, Pintor-Toro J A, Llobell A. Eur J Biochem. 1992;206:859–867. doi: 10.1111/j.1432-1033.1992.tb16994.x. [DOI] [PubMed] [Google Scholar]
- 29.Nagel R J, Manners J M, Birch R G. Plant Mol Biol Report. 1992;10:263–272. [Google Scholar]
- 30.Harman G E, Hayes C K, Lorito M, Broadway R M, Di Pietro A, Tronsmo A. Phytopathology. 1993;83:313–318. [Google Scholar]
- 31.Kuranda M J, Robbins P W. J Biol Chem. 1991;266:19758–19767. [PubMed] [Google Scholar]
- 32.Lawrence C B, Joosten M H A J, Tuzun S. Physiol Mol Plant Pathol. 1996;48:361–377. [Google Scholar]
- 33.Abler M L, Green P J. Plant Mol Biol. 1996;32:63–78. doi: 10.1007/BF00039377. [DOI] [PubMed] [Google Scholar]
- 34.Koziel G M, Carozzi N B, Desai N. Plant Mol Biol. 1996;32:393–405. doi: 10.1007/BF00039392. [DOI] [PubMed] [Google Scholar]
- 35.Collinge D B, Kragh K M, Mikkelsen J D, Nielsen K, Rasmussen U, Vad K. Plant J. 1993;3:31–40. doi: 10.1046/j.1365-313x.1993.t01-1-00999.x. [DOI] [PubMed] [Google Scholar]