Abstract
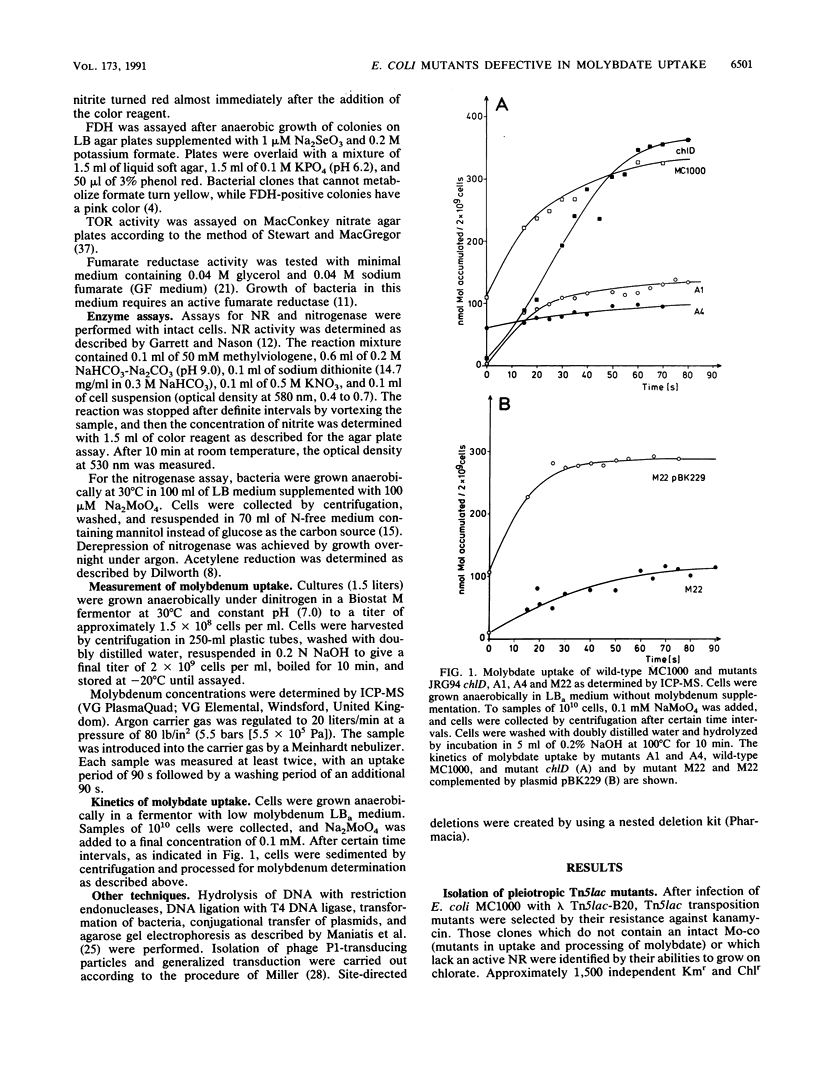
For the study of molybdenum uptake by Escherichia coli, we generated Tn5lac transposition mutants, which were screened for the pleiotropic loss of molybdoenzyme activities. Three mutants A1, A4, and M22 were finally selected for further analysis. Even in the presence of 100 microM molybdate in the growth medium, no active nitrate reductase, formate dehydrogenase, and trimethylamine-N-oxide reductase were detected in these mutants, indicating that the intracellular supply of molybdenum was not sufficient. This was also supported by the observation that introduction of plasmid pWK225 carrying the complete nif regulon of Klebsiella pneumoniae did not lead to a functional expression of nitrogenase. Finally, molybdenum determination by induced coupled plasma mass spectroscopy confirmed a significant reduction of cell-bound molybdenum in the mutants compared with that in wild-type E. coli, even at high molybdate concentrations in the medium. A genomic library established with the plasmid mini-F-derived cop(ts) vector pJE258 allowed the isolation of cosmid pBK229 complementing the molybdate uptake deficiency of the chlD mutant and the Tn5lac-induced mutants. Certain subfragments of pBK229 which do not contain the chlD gene are still able to complement the Tn5lac mutants. Mapping experiments showed that the Tn5lac insertions did not occur within the chromosomal region present in pBK229 but did occur very close to that region. We assume that the Tn5lac insertions have a polar effect, thus preventing the expression of transport genes, or that a positively acting regulatory element was inactivated.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amy N. K., Rajagopalan K. V. Characterization of molybdenum cofactor from Escherichia coli. J Bacteriol. 1979 Oct;140(1):114–124. doi: 10.1128/jb.140.1.114-124.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann B. J. Linkage map of Escherichia coli K-12, edition 7. Microbiol Rev. 1983 Jun;47(2):180–230. doi: 10.1128/mr.47.2.180-230.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann B. J. Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol Rev. 1972 Dec;36(4):525–557. doi: 10.1128/br.36.4.525-557.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F. Construction and characterization of new cloning vehicles. III. Derivatives of plasmid pBR322 carrying unique Eco RI sites for selection of Eco RI generated recombinant DNA molecules. Gene. 1978 Oct;4(2):121–136. doi: 10.1016/0378-1119(78)90025-2. [DOI] [PubMed] [Google Scholar]
- Chippaux M., Giudici D., Abou-Jaoudé A., Casse F., Pascal M. C. Laboratoire de Chimie Bactérienne C.N.R.S., Marsielle, France. Mol Gen Genet. 1978 Apr 6;160(2):225–229. doi: 10.1007/BF00267485. [DOI] [PubMed] [Google Scholar]
- Collins J., Brüning H. J. Plasmids useable as gene-cloning vectors in an in vitro packaging by coliphage lambda: "cosmids". Gene. 1978 Oct;4(2):85–107. doi: 10.1016/0378-1119(78)90023-9. [DOI] [PubMed] [Google Scholar]
- Dilworth M. J. Acetylene reduction by nitrogen-fixing preparations from Clostridium pasteurianum. Biochim Biophys Acta. 1966 Oct 31;127(2):285–294. doi: 10.1016/0304-4165(66)90383-7. [DOI] [PubMed] [Google Scholar]
- Frey B., Jänel G., Michelsen U., Kersten H. Mutations in the Escherichia coli fnr and tgt genes: control of molybdate reductase activity and the cytochrome d complex by fnr. J Bacteriol. 1989 Mar;171(3):1524–1530. doi: 10.1128/jb.171.3.1524-1530.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett R. H., Nason A. Further purification and properties of Neurospora nitrate reductase. J Biol Chem. 1969 Jun 10;244(11):2870–2882. [PubMed] [Google Scholar]
- Glaser J. H., DeMoss J. A. Comparison of nitrate reductase mutants of Escherichia coli selected by alternative procedures. Mol Gen Genet. 1972;116(1):1–10. doi: 10.1007/BF00334254. [DOI] [PubMed] [Google Scholar]
- Glaser J. H., DeMoss J. A. Phenotypic restoration by molybdate of nitrate reductase activity in chlD mutants of Escherichia coli. J Bacteriol. 1971 Nov;108(2):854–860. doi: 10.1128/jb.108.2.854-860.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helsberg M., Ebbers J., Eichenlaub R. Mutations affecting replication and copy number control in plasmid mini-F both reside in the gene for the 29-kDa protein. Plasmid. 1985 Jul;14(1):53–63. doi: 10.1016/0147-619x(85)90032-0. [DOI] [PubMed] [Google Scholar]
- Hohn B., Murray K. Packaging recombinant DNA molecules into bacteriophage particles in vitro. Proc Natl Acad Sci U S A. 1977 Aug;74(8):3259–3263. doi: 10.1073/pnas.74.8.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johann S., Hinton S. M. Cloning and nucleotide sequence of the chlD locus. J Bacteriol. 1987 May;169(5):1911–1916. doi: 10.1128/jb.169.5.1911-1916.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J. L., Hainline B. E., Rajagopalan K. V. Characterization of the molybdenum cofactor of sulfite oxidase, xanthine, oxidase, and nitrate reductase. Identification of a pteridine as a structural component. J Biol Chem. 1980 Mar 10;255(5):1783–1786. [PubMed] [Google Scholar]
- Kennedy C., Postgate J. R. Expression of Klebsiella pneumoniae nitrogen fixation genes in nitrate reductase mutants of Escherichia coli. J Gen Microbiol. 1977 Feb;98(2):551–557. doi: 10.1099/00221287-98-2-551. [DOI] [PubMed] [Google Scholar]
- Lambden P. R., Guest J. R. Mutants of Escherichia coli K12 unable to use fumarate as an anaerobic electron acceptor. J Gen Microbiol. 1976 Dec;97(2):145–160. doi: 10.1099/00221287-97-2-145. [DOI] [PubMed] [Google Scholar]
- Lee J. H., Wendt J. C., Shanmugam K. T. Identification of a new gene, molR, essential for utilization of molybdate by Escherichia coli. J Bacteriol. 1990 Apr;172(4):2079–2087. doi: 10.1128/jb.172.4.2079-2087.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S. F., DeMoss J. A. Promoter region of the nar operon of Escherichia coli: nucleotide sequence and transcription initiation signals. J Bacteriol. 1987 Oct;169(10):4614–4620. doi: 10.1128/jb.169.10.4614-4620.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGregor C. H. Synthesis of nitrate reductase components in chlorate-resistant mutants of Escherichia coli. J Bacteriol. 1975 Mar;121(3):1117–1121. doi: 10.1128/jb.121.3.1117-1121.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. B., Amy N. K. Molybdenum cofactor in chlorate-resistant and nitrate reductase-deficient insertion mutants of Escherichia coli. J Bacteriol. 1983 Aug;155(2):793–801. doi: 10.1128/jb.155.2.793-801.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. B., Scott D. J., Amy N. K. Molybdenum-sensitive transcriptional regulation of the chlD locus of Escherichia coli. J Bacteriol. 1987 May;169(5):1853–1860. doi: 10.1128/jb.169.5.1853-1860.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascal M. C., Burini J. F., Chippaux M. Regulation of the trimethylamine N-oxide (TMAO) reductase in Escherichia coli: analysis of tor::Mud1 operon fusion. Mol Gen Genet. 1984;195(1-2):351–355. doi: 10.1007/BF00332770. [DOI] [PubMed] [Google Scholar]
- Reiss J., Kleinhofs A., Klingmüller W. Cloning of seven differently complementing DNA fragments with chl functions from Escherichia coli K12. Mol Gen Genet. 1987 Feb;206(2):352–355. doi: 10.1007/BF00333594. [DOI] [PubMed] [Google Scholar]
- Scott D., Amy N. K. Molybdenum accumulation in chlD mutants of Escherichia coli. J Bacteriol. 1989 Mar;171(3):1284–1287. doi: 10.1128/jb.171.3.1284-1287.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott J. R. Immunity and repression in bacteriophages P1 and P7. Curr Top Microbiol Immunol. 1980;90:49–65. doi: 10.1007/978-3-642-67717-5_3. [DOI] [PubMed] [Google Scholar]
- Simon R., O'Connell M., Labes M., Pühler A. Plasmid vectors for the genetic analysis and manipulation of rhizobia and other gram-negative bacteria. Methods Enzymol. 1986;118:640–659. doi: 10.1016/0076-6879(86)18106-7. [DOI] [PubMed] [Google Scholar]
- Simon R., Quandt J., Klipp W. New derivatives of transposon Tn5 suitable for mobilization of replicons, generation of operon fusions and induction of genes in gram-negative bacteria. Gene. 1989 Aug 1;80(1):161–169. doi: 10.1016/0378-1119(89)90262-x. [DOI] [PubMed] [Google Scholar]
- Sperl G. T., DeMoss J. A. chlD gene function in molybdate activation of nitrate reductase. J Bacteriol. 1975 Jun;122(3):1230–1238. doi: 10.1128/jb.122.3.1230-1238.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart V., MacGregor C. H. Nitrate reductase in Escherichia coli K-12: involvement of chlC, chlE, and chlG loci. J Bacteriol. 1982 Aug;151(2):788–799. doi: 10.1128/jb.151.2.788-799.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. L., Bedbrook J. R., Grant F. J., Kleinhofs A. Reconstitution of plant nitrate reductase by Escherichia coli extracts and the molecular cloning of the chlA gene of Escherichia coli K12. J Mol Appl Genet. 1983;2(3):261–271. [PubMed] [Google Scholar]
- Venables W. A., Guest J. R. Transduction of nitrate reductase loci of Escherichia coli by phages P-1 and lambda. Mol Gen Genet. 1968;103(2):127–140. doi: 10.1007/BF00427140. [DOI] [PubMed] [Google Scholar]



