Abstract
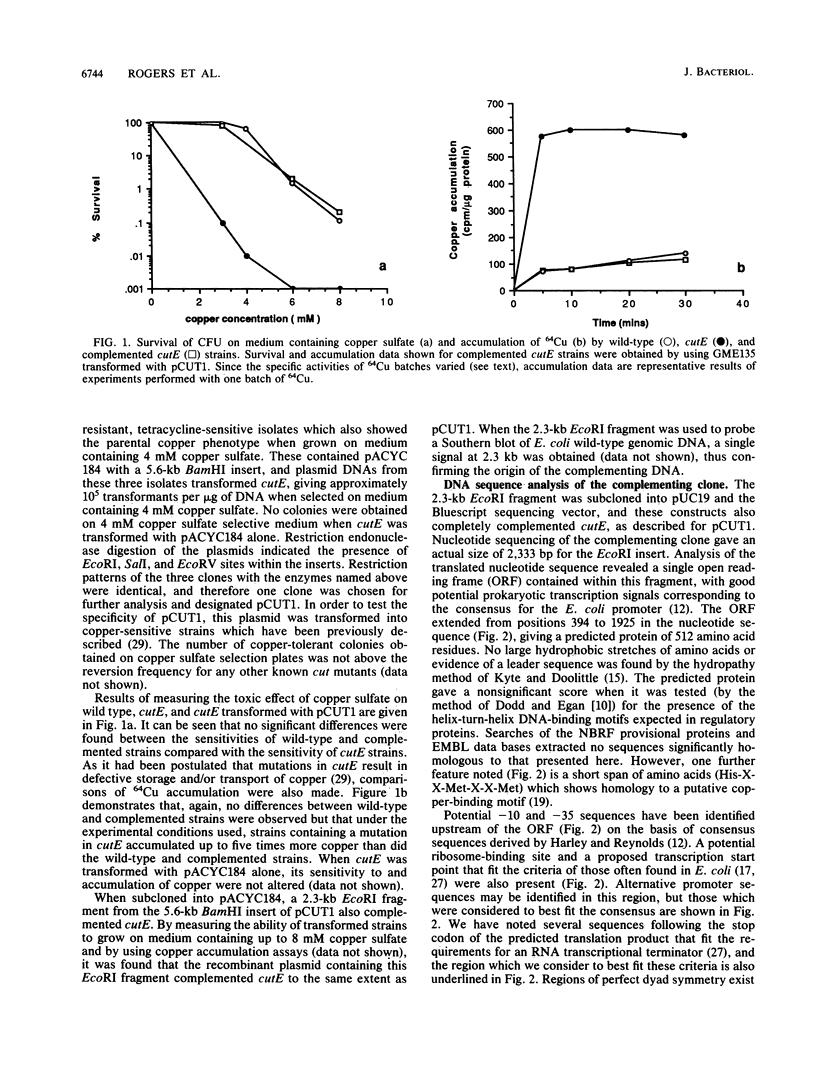
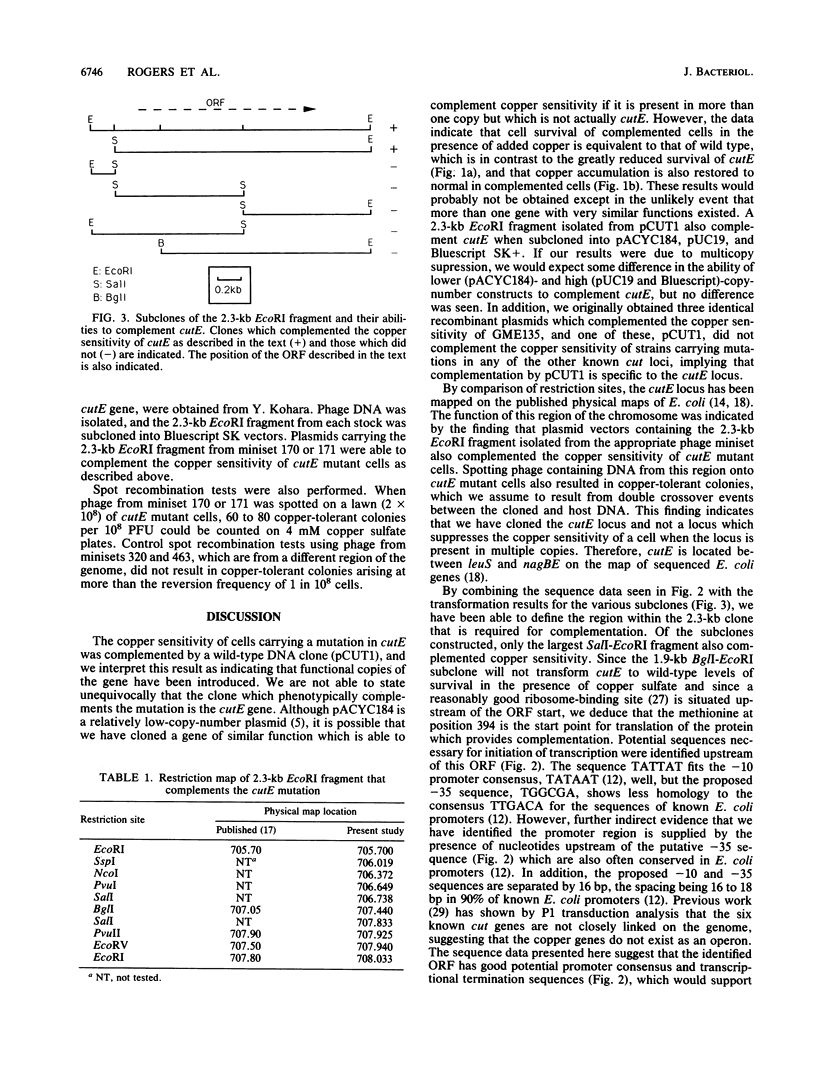
The copper-sensitive/temperature-sensitive phenotype of the Escherichia coli cutE mutant has been complemented by cloning wild-type genomic DNA into the plasmid vector pACYC184 and selecting transformants on medium containing 4 mM copper sulfate and chloramphenicol. One of these complementing clones, designated pCUT1, contained a 5.6-kb BamHI fragment. This recombinant plasmid transformed cutE, allowing wild-type growth of transformants on medium containing copper sulfate. Complementation of copper sensitivity was assessed by comparing both cell survival at increased copper levels and the results of 64Cu accumulation assays. An EcoRI subclone, 2.3 kb in size, was also shown to complement cutE when cloned in both medium- and high-copy-number vectors and was completely sequenced. This clone was mapped on the E. coli physical map at 705.70 to 707.80 kb. A series of subclones was constructed from pCUT1 and used to show that the large open reading frame of the translated sequence was essential for complementation. This open reading frame has a potential upstream promoter region, ribosome-binding site, and transcriptional terminator and encodes a putative protein of 512 amino acids that contains a region showing some homology to a putative copper-binding site.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Borck K., Beggs J. D., Brammar W. J., Hopkins A. S., Murray N. E. The construction in vitro of transducing derivatives of phage lambda. Mol Gen Genet. 1976 Jul 23;146(2):199–207. doi: 10.1007/BF00268089. [DOI] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosa J. H. Genetics and molecular biology of siderophore-mediated iron transport in bacteria. Microbiol Rev. 1989 Dec;53(4):517–530. doi: 10.1128/mr.53.4.517-530.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd I. B., Egan J. B. Improved detection of helix-turn-helix DNA-binding motifs in protein sequences. Nucleic Acids Res. 1990 Sep 11;18(17):5019–5026. doi: 10.1093/nar/18.17.5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutteridge J. M., Wilkins S. Copper salt-dependent hydroxyl radical formation. Damage to proteins acting as antioxidants. Biochim Biophys Acta. 1983 Aug 23;759(1-2):38–41. doi: 10.1016/0304-4165(83)90186-1. [DOI] [PubMed] [Google Scholar]
- Harley C. B., Reynolds R. P. Analysis of E. coli promoter sequences. Nucleic Acids Res. 1987 Mar 11;15(5):2343–2361. doi: 10.1093/nar/15.5.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin M. Metallothioneins: proteins in search of function. Cell. 1985 May;41(1):9–10. doi: 10.1016/0092-8674(85)90051-0. [DOI] [PubMed] [Google Scholar]
- Kohara Y., Akiyama K., Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987 Jul 31;50(3):495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Mason H. S. Binuclear copper clusters as active sites for oxidases. Adv Exp Med Biol. 1976;74:464–469. doi: 10.1007/978-1-4684-3270-1_39. [DOI] [PubMed] [Google Scholar]
- McClure W. R. Mechanism and control of transcription initiation in prokaryotes. Annu Rev Biochem. 1985;54:171–204. doi: 10.1146/annurev.bi.54.070185.001131. [DOI] [PubMed] [Google Scholar]
- Mellano M. A., Cooksey D. A. Nucleotide sequence and organization of copper resistance genes from Pseudomonas syringae pv. tomato. J Bacteriol. 1988 Jun;170(6):2879–2883. doi: 10.1128/jb.170.6.2879-2883.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra T. K., Brown N. L., Fritzinger D. C., Pridmore R. D., Barnes W. M., Haberstroh L., Silver S. Mercuric ion-resistance operons of plasmid R100 and transposon Tn501: the beginning of the operon including the regulatory region and the first two structural genes. Proc Natl Acad Sci U S A. 1984 Oct;81(19):5975–5979. doi: 10.1073/pnas.81.19.5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Médigue C., Bouché J. P., Hénaut A., Danchin A. Mapping of sequenced genes (700 kbp) in the restriction map of the Escherichia coli chromosome. Mol Microbiol. 1990 Feb;4(2):169–187. doi: 10.1111/j.1365-2958.1990.tb00585.x. [DOI] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- O'Halloran T. V., Frantz B., Shin M. K., Ralston D. M., Wright J. G. The MerR heavy metal receptor mediates positive activation in a topologically novel transcription complex. Cell. 1989 Jan 13;56(1):119–129. doi: 10.1016/0092-8674(89)90990-2. [DOI] [PubMed] [Google Scholar]
- O'Halloran T., Walsh C. Metalloregulatory DNA-binding protein encoded by the merR gene: isolation and characterization. Science. 1987 Jan 9;235(4785):211–214. doi: 10.1126/science.3798107. [DOI] [PubMed] [Google Scholar]
- Olafson R. W., McCubbin W. D., Kay C. M. Primary- and secondary-structural analysis of a unique prokaryotic metallothionein from a Synechococcus sp. cyanobacterium. Biochem J. 1988 May 1;251(3):691–699. doi: 10.1042/bj2510691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouzounis C., Sander C. A structure-derived sequence pattern for the detection of type I copper binding domains in distantly related proteins. FEBS Lett. 1991 Feb 11;279(1):73–78. doi: 10.1016/0014-5793(91)80254-z. [DOI] [PubMed] [Google Scholar]
- Parkhill J., Brown N. L. Site-specific insertion and deletion mutants in the mer promoter-operator region of Tn501; the nineteen base-pair spacer is essential for normal induction of the promoter by MerR. Nucleic Acids Res. 1990 Sep 11;18(17):5157–5162. doi: 10.1093/nar/18.17.5157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Rouch D., Camakaris J., Lee B. T., Luke R. K. Inducible plasmid-mediated copper resistance in Escherichia coli. J Gen Microbiol. 1985 Apr;131(4):939–943. doi: 10.1099/00221287-131-4-939. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short J. M., Fernandez J. M., Sorge J. A., Huse W. D. Lambda ZAP: a bacteriophage lambda expression vector with in vivo excision properties. Nucleic Acids Res. 1988 Aug 11;16(15):7583–7600. doi: 10.1093/nar/16.15.7583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver S., Misra T. K. Plasmid-mediated heavy metal resistances. Annu Rev Microbiol. 1988;42:717–743. doi: 10.1146/annurev.mi.42.100188.003441. [DOI] [PubMed] [Google Scholar]
- Simpson J. A., Cheeseman K. H., Smith S. E., Dean R. T. Free-radical generation by copper ions and hydrogen peroxide. Stimulation by Hepes buffer. Biochem J. 1988 Sep 1;254(2):519–523. doi: 10.1042/bj2540519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers A. O. Organization, expression, and evolution of genes for mercury resistance. Annu Rev Microbiol. 1986;40:607–634. doi: 10.1146/annurev.mi.40.100186.003135. [DOI] [PubMed] [Google Scholar]
- Tappel A. L. Lipid peroxidation damage to cell components. Fed Proc. 1973 Aug;32(8):1870–1874. [PubMed] [Google Scholar]
- Tetaz T. J., Luke R. K. Plasmid-controlled resistance to copper in Escherichia coli. J Bacteriol. 1983 Jun;154(3):1263–1268. doi: 10.1128/jb.154.3.1263-1268.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Moore M., Levinson H. S., Silver S., Walsh C., Mahler I. Nucleotide sequence of a chromosomal mercury resistance determinant from a Bacillus sp. with broad-spectrum mercury resistance. J Bacteriol. 1989 Jan;171(1):83–92. doi: 10.1128/jb.171.1.83-92.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lorenzo V., Wee S., Herrero M., Neilands J. B. Operator sequences of the aerobactin operon of plasmid ColV-K30 binding the ferric uptake regulation (fur) repressor. J Bacteriol. 1987 Jun;169(6):2624–2630. doi: 10.1128/jb.169.6.2624-2630.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]


