Abstract
Cooperative DNA binding by transcription factors that bind to separate recognition sites is likely to require bending of intervening sequences and the appropriate orientation of transcription factor binding. We investigated DNA bending in complexes formed by the basic region–leucine zipper domains of Fos and Jun with the DNA binding region of nuclear factor of activated T cells 1 (NFAT1) at composite regulatory elements using gel electrophoretic phasing analysis. The NFAT1—Fos–Jun complex induced a bend at the ARRE2 site that was distinct from the sum of the bends induced by NFAT1 and Fos–Jun separately. We designate this difference DNA bending cooperativity. The bending cooperativity was directed toward the interaction interface between Fos–Jun and NFAT1. We also examined the influence of NFAT1 on the orientation of Fos–Jun heterodimer binding using a novel fluorescence resonance energy transfer assay. The interaction with NFAT1 could reverse the orientation of Fos–Jun heterodimer binding to the ARRE2 site. The principal determinants of both cooperative DNA bending and oriented heterodimer binding were localized to three amino acid residues at the amino-terminal ends of the leucine zippers of Fos and Jun. Consequently, interactions between transcription factors can remodel promoters by altering DNA bending and the orientation of heterodimer binding.
Transcription of most genes in eukaryotic organisms is regulated through the cooperative action of multiple transcription regulatory proteins that bind to separate DNA sequence elements. One mechanism of transcriptional cooperativity is the stabilization of transcription factor binding through interactions among proteins that bind to different DNA recognition sites (1–4). Interactions between proteins that bind to separate promoter elements are likely to require DNA bending and the correct orientation of transcription factor binding to allow juxtaposition of the molecular surfaces that mediate the interaction.
One example of transcriptional cooperativity is observed in complexes formed by nuclear factor of activated T cells (NFAT) family proteins with Fos and Jun families of basic region–leucine zipper (bZIP) proteins. Members of the NFAT and the Fos–Jun families bind cooperatively to adjoining NFAT and AP-1 recognition sequences and regulate cytokine gene expression in activated T cells (1, 2). The composite regulatory elements that mediate cytokine gene activation in response to antigen stimulation generally contain non-consensus recognition sequences for NFAT1 and Fos–Jun and bind weakly to either component of the complex alone (1). Cooperative binding by NFAT1 and Fos–Jun stabilizes the complex and promotes synergistic transcription activation (2).
The effects of individual transcription factor complexes on DNA structure have been extensively investigated. Fos and Jun induce opposite directions of DNA bending based on gel electrophoretic phasing analysis (5, 6). The opposite directions of DNA bending are caused by the converse electrostatic interactions between Fos and Jun and the phosphodiester backbone (7–12). No significant DNA bending was observed in the x-ray crystal structure of the bZIP domains of Fos and Jun or in cyclization or minicircle binding experiments (13, 14). The crystallization conditions and the stacking of oligonucleotides in adjacent unit cells may constrain the conformational flexibility of the binding site and prevent bending in the crystal (7). The truncated Fos and Jun proteins used in the cyclization and minicircle binding assays induce DNA bends of similar magnitudes in opposite directions, resulting in a net bend that is smaller than the detection limit of these assays (14–16). The effects of multiprotein complexes on DNA structure have been examined in a few cases (17–20), but the relative roles of the individual proteins and by their interactions have not been defined. We have examined the effects of interactions between Fos–Jun heterodimers and NFAT1 on DNA bending and on the orientation of Fos–Jun heterodimer binding to DNA.
MATERIALS AND METHODS
Plasmid Construction and Protein Purification.
Plasmids containing the ARRE2 (AAAGAGGAAAATTTGTTTCATAC) (pNR710), ARRE2(AP-1) (AAAGAGGAAAATTTGACTCATAC) (pNR711), and CD28RE (AAAGAAATTCCAGAGAGTCATCA) (pNR720) sites at different distances from an intrinsic reference bend were constructed by ligation of duplex oligonucleotides between the unique XbaI and SalI sites of plasmids pTK401–26 and pTK401–28 as described (9). The plasmids used for expression of proteins encompassing the bZIP domains of Fos and Jun and the DNA binding region of NFAT1 (residues 396–692) have been described (1, 7, 16, 21). Plasmids encoding the bZIP domains of Fos and Jun with single Cys residues at different positions on the amino-terminal side of the basic regions were constructed by substitution of Cys-154 in Fos and Cys-272 in Jun with Ser and residue 132, 136, or 142 of Fos and residue 251, 253, or 260 of Jun with Cys. Plasmids encoding chimeric proteins were constructed by PCR amplification of the appropriate regions of these contructs. All proteins were purified by nickel chelate affinity chromatography to greater than 90% homogeneity (5).
Phasing Analysis and Quantitation of Bending.
Probes for phasing analysis were prepared by PCR amplification as described (5) using plasmids pNR710-n, pNR711-n, and pNR720-n as templates (n = 26, 28, 30, 32, 34, or 36). The electrophoretic mobilities of the complexes were normalized for differences in the mobilities of the probes alone to determine protein-induced DNA bending (7, 9). The average and standard deviation of results from three or more independent experiments are shown. The magnitudes and directions of the DNA bends were calculated as described (16).
Two methods were used to determine the bending cooperativity. First, the combined effect of the DNA bends induced by the individual proteins was obtained by adding vectors representing the magnitudes and directions of the bends (6, 16). The bending cooperativity was calculated by subtracting the vector representing the combined effect of the DNA bends induced by the individual subunits from a vector representing the bend induced by the complex. Second, the combined effect of the mobility variations caused by the individual complexes was calculated based on the relationship between DNA bend angle and electrophoretic mobility (8). The contribution of bending cooperativity to complex mobilities was calculated from the ratio between the observed mobilities and the mobilities predicted based on the combined effect of the individual bends. Bending cooperativity was invoked only when the combined effect of the individual bends deviated significantly (P < 0.001) from the bend induced by the intact complex based on both methods.
Fluorescence Architectural Analysis.
Oligonucleotides 27 bp in length containing the ARRE2, ARRE2(AP-1), or CD28RE site with the AP-1 recognition sequence at the center were labeled on one end by using fluorescein phosphoramidite (Glen Research, Sterling, VA). The three base pairs at both ends of each oligonucleotide were made identical to standardize the local environment of the donor fluorophores. The annealed duplexes were purified by PAGE to remove incomplete synthesis products and duplexes lacking the fluorophore. Proteins encompassing the bZIP domains of Fos and Jun with unique Cys residues were reacted with Texas Red C2 maleimide or X-rhodamine-5/6-iodoacetamide (Molecular Probes) and purified as described (22). The purified proteins were analyzed by SDS/PAGE and were confirmed to be greater than 90% labeled based on the shift in the electrophoretic mobility of labeled proteins. Complexes formed by heterodimers on the labeled oligonucleotides were separated by PAGE, and the donor and acceptor fluorescence was quantitated by scanning the gel in a FluorImager (Molecular Dynamics) with excitation at 488 nm. The light emitted at each position in the gel was passed alternately through 530 ± 30 nm bandpass and 610 nm cut-on interference filters. The emission from each fluorophore was calculated by comparing the emission through each filter with the emissions of calibration standards containing pure donor and acceptor fluorophores.
The efficiency of energy transfer is defined here as the ratio between acceptor and donor emission (TR/FL). The end preference was calculated by dividing the efficiency of energy transfer from the left end of the oligonucleotide (L) by the sum of the efficiencies of energy transfer from the left (L) and right (R) ends (L/L+R).
RESULTS
Fos–Jun and NFAT1 Cooperate to Bend DNA.
We have investigated the effects of cooperative DNA binding by Fos, Jun, and NFAT1 on DNA bending and on the orientation of Fos–Jun heterodimer binding at different binding sites. The minimal regions required for cooperative complex formation, the bZIP domains of Fos and Jun and the DNA binding region of NFAT1 (1, 21), were tested. The DNA bends induced by NFAT1 and by Fos–Jun individually as well as by the NFAT1—Fos–Jun complex were compared using gel electrophoretic phasing analysis (Fig. 1A). In this assay, the spacing between a protein–DNA complex and a reference bend is varied, and the magnitude and direction of the protein-induced DNA bend are quantitated based on the variation in electrophoretic mobility (Fig. 1B) (5, 6).
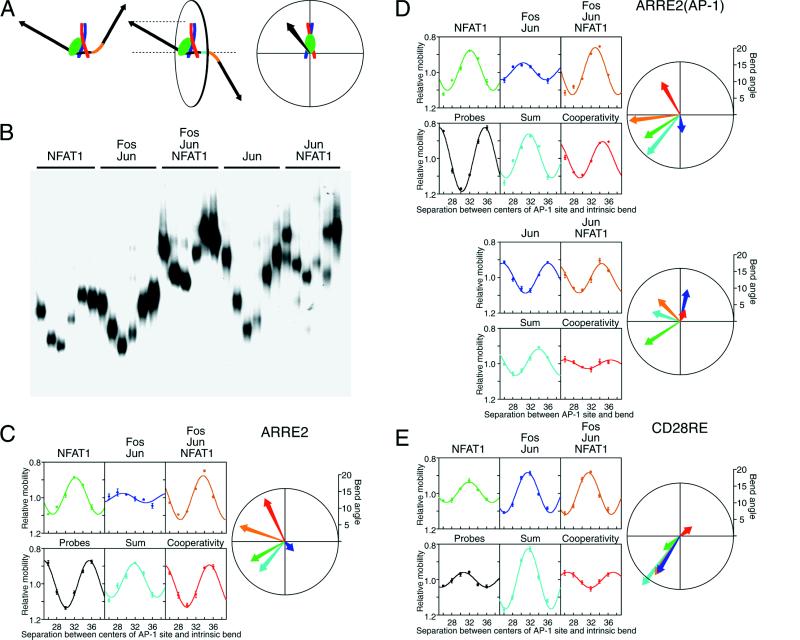
Figure 1.
DNA bending cooperativity in complexes formed by the DNA binding region of NFAT1 and the bZIP domains of Fos and Jun. (A) Schematic representation of phasing analysis and the polar representation of DNA bending. The two diagrams on the left represent NFAT1—Fos–Jun complexes (NFAT1, green; Fos, red; Jun, blue) that differ in the spacing (cyan) between the protein-induced and intrinsic (orange) DNA bends. When the bends are in phase (left complex), the overall bend is larger, and the electrophoretic mobility is lower than when the two bends are out of phase (middle complex). The diagrams on the right describe the polar representation of DNA bending, which is equivalent to the projection of the bent DNA helix onto a plane perpendicular to the DNA helix axis between the AP-1 site and the intrinsic bend. The orientation and length of the vector show the direction and magnitude of the protein-induced DNA bend (16). (B) Phasing analysis of DNA bending by Fos(139–200)–Jun(257–318) heterodimers and Jun(241–334) homodimers in the presence and absence of NFAT1(396–692). The proteins indicated above the lanes were incubated with DNA fragments that differ in the spacing between the ARRE2(AP-1) site and the intrinsic bend (26, 28, 30, 32, 34, and 36 bp), and the complexes were analyzed by PAGE. (C) Quantitation of DNA bending by complexes formed by NFAT1, Fos, and Jun at the ARRE2 site. The mobilities of the complexes were normalized for differences in probe mobilities and plotted as a function of the separation between the centers of the AP-1 recognition sequence and the intrinsic bend. The predicted additive effect of the bends induced by NFAT1 and Fos–Jun heterodimers separately is shown in the graph (cyan) below the Fos–Jun complex mobilities. The ratio between the relative mobilities of the NFAT1—Fos–Jun—ARRE2 complexes and the relative mobilities of complexes formed by NFAT1 and Fos–Jun separately reflects the contribution of bending cooperativity to complex mobility. This effect of bending cooperativity is shown in the graph (red) below the NFAT1—Fos–Jun—ARRE2 complex mobilities. The direction and magnitude of the DNA bend induced by each complex were calculated as described (16) and are shown together with the sum of the bends and the bending cooperativity in polar representation using the same colors that were used to plot their relative mobilities. (D) Quantitation of DNA bending by complexes formed by NFAT1, Fos, and Jun at the ARRE2(AP-1) site. (E) Quantitation of DNA bending by complexes formed by NFAT1, Fos, and Jun at the CD28RE site.
Fos–Jun heterodimers, NFAT1 alone, and the NFAT1—Fos–Jun complex induced DNA bending at the ARRE2 site in the interleukin-2 promoter (Fig. 1C). The DNA bend induced by the NFAT1—Fos–Jun complex (orange) was distinct from the combined effect of the bends induced by Fos–Jun and NFAT1 separately (cyan). Thus, DNA bending by the NFAT1—Fos–Jun complex did not reflect the simple additive effect of independent DNA bends induced by Fos–Jun and NFAT1. This is in contrast to the independent effects of Fos and Jun on DNA bending by the heterodimer (16) as well as the additive effects of closely spaced intrinsic DNA bends (8). The difference between the bend induced by the complex and the sum of the bends induced by the individual components is here designated bending cooperativity (red). We hypothesize that the bending cooperativity in the NFAT1—Fos–Jun complex is caused by an interaction between Fos–Jun and NFAT1 that induces additional DNA bending.
To investigate the molecular basis of the bending cooperativity between Fos–Jun and NFAT1, complexes formed by homo- and heterodimers were examined at several binding sites. The ARRE2 site binds Fos–Jun heterodimers alone with low affinity and does not bind Jun homodimers alone. To test the influence of DNA binding affinity on bending cooperativity and to determine the bending cooperativity between Jun homodimers and NFAT1, DNA bending was examined at the ARRE2(AP-1) site containing a consensus AP-1 recognition sequence (Fig. 1D). The DNA bending cooperativity between Fos–Jun heterodimers and NFAT1 at this site was similar to that observed at the ARRE2 site, indicating that the bending cooperativity was not caused by the low binding affinity of Fos–Jun heterodimers alone at the ARRE2 site.
Jun homodimers also induced DNA bending at the ARRE2(AP-1) site. In contrast to Fos–Jun heterodimers, the DNA bend induced by Jun homodimers together with NFAT1 was nearly identical to the sum of the bends induced by Jun homodimers and NFAT1 separately. Thus, there is a specific interaction between Fos–Jun heterodimers and NFAT1 that is required for bending cooperativity at the ARRE2 site.
To examine the influence of the arrangement of the NFAT and AP-1 recognition sequences on bending cooperativity, DNA bending at the ARRE2 site was compared with bending at the CD28RE site (Fig. 1E). There was little DNA bending cooperativity between Fos–Jun and NFAT1 at the CD28RE site. The weak bending cooperativity was in a direction diametrically opposite to the bends induced by Fos–Jun and NFAT1 and resulted in a slightly smaller overall bend than predicted by the sum of the independent bends. These results indicate that the bending cooperativity between Fos–Jun and NFAT1 was affected by the alignment of the complexes on DNA.
NFAT1 Controls the Orientation of Fos–Jun Heterodimer Binding.
Cooperative DNA binding with NFAT1 causes Fos–Jun to bind to the ARRE2 site in a preferred orientation (23). We have compared the effects of the interaction between Fos–Jun and NFAT1 on DNA bending and on the orientation of heterodimer binding. We have previously shown that Fos–Jun heterodimers bind to AP-1 sites in a preferred orientation and that mutation of a conserved arginine in Fos vis-à-vis Jun causes heterodimers to bind in opposite orientations (7, 9). We tested the effects of these mutations on the orientation of heterodimer binding in the presence and absence of NFAT1 (Fig. 2). A novel assay based on fluorescence resonance energy transfer (FRET) between a donor fluorophore linked to one end of the oligonucleotide and an acceptor fluorophore coupled to one subunit of the heterodimer was used to determine the orientation of heterodimer binding (Fig. 2A). We designate this approach fluorescence architectural analysis.
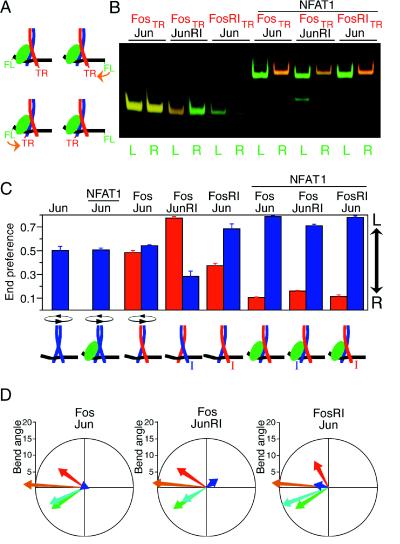
Figure 2.
Influence of asymmetric recognition of the AP-1 site and the interaction with NFAT1 on the orientation of Fos–Jun heterodimer binding. (A) Schematic representation of the determination of the orientation of heterodimer binding by FRET. The efficiency of energy transfer (orange arrow) is higher when the donor fluorescein (FL) and the acceptor Texas Red (TR) are on the same side of the binding site. (B) FRET analysis of the influence of asymmetric recognition of the AP-1 site on the orientation of heterodimer binding in the absence and presence of NFAT1. Heterodimers formed between Fos(139–200) labeled with Texas Red (FosTR) and Jun(257–318) (Jun) as well as heterodimers formed by Fos(139–200)R155I (FosRI) and Jun(257–318)R273I (JunRI) with the wild-type proteins were incubated with oligonucleotides labeled at the 5′-end with fluorescein on either the left (L) or the right (R) side of the ARRE2(AP-1) site. Complexes were formed in the presence or the absence of NFAT1(396–692) as indicated. The complexes were separated by PAGE, and the gel was scanned using a laser that excites the fluorescein donor. The donor (green) and acceptor (red) emissions at each position in the gel were quantitated, and the pseudocolor images were superimposed. Thus, different ratios of donor and acceptor fluorescence result in bands of different colors. (C) Quantitation of the relative efficiencies of energy transfer from opposite ends of the oligonucleotide. The end preference reflects the ratio between the efficiencies of energy transfer from opposite ends of the oligonucleotide. A high end preference indicates a higher efficiency of energy transfer from the left end, whereas a low end preference indicates a higher efficiency of energy transfer from the right end (indicated by the arrow on the right). Complexes containing labeled Fos are shown by the red bars, and complexes containing labeled Jun are shown by the blue bars. The end preferences of Jun homodimers in the absence and presence of NFAT1 are shown on the left. The inferred orientation of heterodimer binding in each complex is shown at the bottom of the figure. The I indicates the basic region containing a mutation in the conserved arginine (7). (D) Influence of the orientation of heterodimer binding on bending cooperativity. DNA bending by heterodimers formed among Fos(118–211) (Fos), Jun(225–334) (Jun), Fos(118–211)R155I (FosRI), and Jun(225–334)R273I (JunRI) (7) in the presence and absence of NFAT1(396–692) was analyzed at the ARRE2(AP-1) site. The DNA bends induced by the complexes were calculated and plotted using the same colors as in Fig. 1.
The relative efficiencies of energy transfer from opposite ends of the oligonucleotide were quantitated in parallel by measuring donor (green) and acceptor (red) emission in complexes separated by PAGE (Fig. 2B). Heterodimers formed by wild-type Fos and Jun bound to this site in both orientations in the absence of NFAT1. Heterodimers in which the conserved arginine of one or the other subunit was mutated bound to the site in opposite orientations (compare FosTR–JunRI and FosRITR–Jun). In the presence of NFAT1, all heterodimers bound to the ARRE2(AP-1) and ARRE2 sites in the same orientation. Thus, the interaction with NFAT1 controlled the orientation of heterodimer binding at these sites. There was no significant effect of NFAT1 on the relative efficiencies of energy transfer at the CD28RE site (data not shown). Therefore, these sites differ in both the effect of NFAT1 on the orientation of Fos–Jun heterodimer binding as well as in bending cooperativity.
Determination of the efficiencies of energy transfer from opposite ends of the oligonucleotide to each subunit provides a quantitative measure of the orientation preference of heterodimer binding (Fig. 2C). Differences in end preference are predicted to be directly proportional to differences in the fraction of heterodimers bound in each orientation. The absolute orientation preference can only be determined by quantitation of the efficiencies of energy transfer for complexes that bind in a unique orientation. Heterodimers containing labeled Fos (red bars) and the same heterodimer containing labeled Jun (blue bars) exhibited complementary end preferences. To exclude the possibility that fluorophore modification of the proteins affected their properties, both Fos and Jun were labeled at three separate positions using Texas Red or rhodamine. The position and identity of the fluorophore did not alter the orientation of heterodimer binding in the presence or absence of NFAT1.
We have previously shown that the orientation of heterodimer binding affects DNA bending by Fos and Jun (7, 9). To determine if the shift in the orientation of heterodimer binding in the presence of NFAT1 contributed to the bending cooperativity, we compared heterodimers whose orientation was reversed in the presence of NFAT1 (Fos–JunRI) with heterodimers whose orientation was less affected (FosRI–Jun) (Fig. 2D). There was a slight difference in the direction of the bending cooperativity between these heterodimers, but the magnitude of the bending cooperativity was similar for all complexes. Thus, the shift in the orientation of heterodimer binding in the presence of NFAT1 was not the principal cause of cooperative DNA bending in the NFAT1—Fos–Jun—ARRE2 complex.
Residues in Fos and Jun That Determine the Orientation of Heterodimer Binding and Cooperative DNA Bending with NFAT1.
To determine the relationship between the effects of Fos–Jun interaction with NFAT1 on the orientation of heterodimer binding and cooperative DNA bending, we sought to identify the residues in Fos and Jun that influence these phenomena. Chimeric proteins were prepared in which successively shorter segments were exchanged between the bZIP domains of Fos and Jun (Fig. 3A). The orientations of heterodimers formed by the chimeric proteins in the presence of NFAT1 were determined using FRET (Fig. 3B). Exchange of segments encompassing the entire leucine zippers between Fos and Jun (J1F–F1J, J2F–F2J) reversed the orientation of basic region binding relative to the wild-type proteins (Fig. 3C). In contrast, exchange of the leucine zippers beginning with the fourth residue after the first leucine (J3F–F3J) did not alter the orientation of basic region binding compared with wild-type Fos and Jun. There was no effect of exchange of these regions on the orientation of heterodimer binding in the absence of NFAT1. These results indicate that the three amino acid residues following the first leucine in the leucine zippers of Fos and Jun comprise the principal determinant of the asymmetric interaction between Fos–Jun and NFAT1.
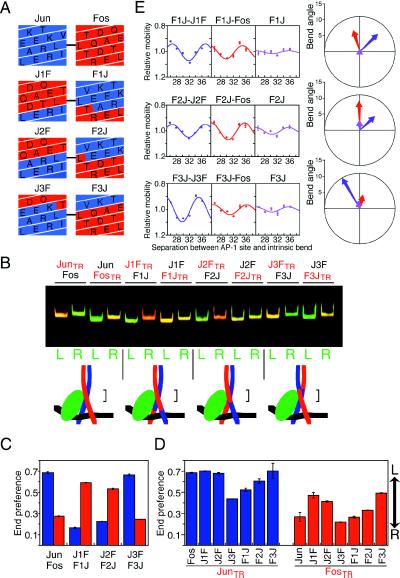
Figure 3.
Localization of amino acid residues in Fos and Jun that influence the orientation of heterodimer binding and bending cooperativity. (A) Helical net diagrams of the amino-terminal ends of the leucine zipper and spacer regions of wild-type Jun (blue) and Fos (red) and chimeric proteins in which progressively shorter segments of the bZIP regions were exchanged (J1F and F1J; J2F and F2J; J3F and F3J). The designations of the chimeric proteins reflect the origins of the amino- and carboxyl-terminal ends of the bZIP domains (F, Fos; J, Jun) and the positions of the junctions. The first leucines in the zipper of each heterodimer are connected by a line. The distal part of the leucine zipper is above and the basic region is below the region depicted. The coiled coil is unwound and viewed from the side occupied by NFAT1 (the same perspective as the diagram on the right side of Fig. 1A). (B) Heterodimers formed between wild-type Fos(134–200) (Fos) and Jun(252–318) (Jun) as well as the chimeric proteins indicated above the lanes (the subunits shown in red were labeled with Texas Red) and NFAT1(396–692) were incubated with oligonucleotides labeled at the 3′-end with fluorescein on either the left (L) or the right (R) side of the ARRE2(AP-1) site, and the complexes were analyzed as described in Fig. 2B. The diagrams below the gel indicate the inferred orientation of heterodimer binding. The bracket to the right of each complex indicates the region that determines the orientation of heterodimer binding. (C) The end preferences for heterodimers formed by the chimeric proteins on oligonucleotides containing the ARRE2(AP-1) site labeled on the 3′-ends were quantitated. Complexes labeled on the amino-terminal end of the basic region from Fos and from Jun are shown in red and blue bars, respectively. Identical results were obtained for complexes bound to the ARRE2 site. (D) The end preference of heterodimers formed between the chimeric proteins and the bZIP domains of wild-type Fos and Jun on oligonucleotides containing the ARRE2(AP-1) site, labeled on the 5′-ends. Complexes labeled on Jun are shown by blue bars, and complexes labeled on Fos are shown by red bars. (E) Bending cooperativity of homo- and heterodimers formed by the chimeric proteins with NFAT1(396–692) at the ARRE2(AP-1) site. Each set of complexes contains the same chimeric protein in association with different dimerization partners. The contribution of bending cooperativity to the mobility variation of the complexes indicated above the graphs was determined as described in Fig. 1. The bending cooperativity is shown in polar representation using different colors to indicate the different complexes.
To evaluate the contributions of other regions of the bZIP domains to the asymmetric interaction between Fos–Jun and NFAT1, we determined the orientations of heterodimers formed between the chimeric proteins and wild-type Fos and Jun in the presence of NFAT1 (Fig. 3D). Heterodimers in which the three residues at the amino-terminal ends of the leucine zippers were derived from Fos and Jun in the two subunits (Jun–J1F, Jun–J2F, Jun–F3J, Fos–J3F, Fos–F1J, and Fos–F2J) bound in the same orientation as the wild-type proteins. However, heterodimers in which these residues were identical in both subunits (Jun–J3F, Jun–F1J, Jun–F2J, Fos–J1F, Fos–J2F, and Fos–F3J) exhibited reduced orientation preference. There was a small effect of the two amino acid residues preceding the first leucine in the leucine zippers on the orientation of heterodimer binding, but the exchange of other segments within the basic or spacer regions or in the distal part of the leucine zippers did not significantly alter the orientation preference. Thus, regardless of the positions of other regions in the heterodimer, the subunit containing the amino-terminal end of the leucine zipper of Jun bound to the half-site closer to NFAT1, whereas the subunit containing this region from Fos bound to the distal half-site in the presence of NFAT1.
To test whether the residues that determine the orientation of heterodimer binding also influence bending cooperativity, we examined DNA bending by complexes formed by the chimeric proteins (Fig. 3E). All heterodimers formed between chimeric proteins generated through a reciprocal exchange of sequences between Fos and Jun exhibited cooperative bending with NFAT1. The direction of the cooperative bend induced by heterodimers in which the complete leucine zipper had been exchanged (F1J–J1F and F2J–J2F) was distinct from that of heterodimers in which the leucine zipper beginning with the fourth residue after the first leucine had been exchanged (F3J–J3F). This difference is consistent with the opposite orientations of binding by these heterodimers in the presence of NFAT1 (Fig. 3C). In contrast, homodimers formed by the chimeric proteins containing the basic region of Fos and the leucine zipper of Jun exhibited little bending cooperativity. These results indicate that residues from the same region of the leucine zippers of both Fos and Jun are required for bending cooperativity with NFAT1.
To identify the residues required for bending cooperativity, heterodimers formed between the chimeric proteins and wild-type Fos were examined. Heterodimers formed by chimeric proteins containing the complete leucine zipper from Jun with wild-type Fos (F1J–Fos and F2J–Fos) exhibited a bending cooperativity that was similar in magnitude to that of heterodimers formed between the respective chimeric proteins (F1J–J1F and F2J–J2F). In contrast, heterodimers formed by chimeric proteins containing the first three residues from the leucine zipper of Fos with wild-type Fos (F3J–Fos) exhibited a small bending cooperativity similar to that of homodimers formed by the chimeric proteins (F3J). Heterodimers formed between the chimeric proteins and wild-type Fos have symmetrical basic regions and are therefore predicted to bend DNA in a direction parallel to the leucine zipper axis regardless of the orientation of heterodimer binding. Thus, in perfect agreement with the determinants of the orientation of heterodimer binding, heterodimers in which the residues at the amino-terminal ends of the leucine zippers were derived from Fos and Jun exhibited cooperative DNA bending with NFAT1, whereas heterodimers in which these residues were identical in both subunits exhibited little bending cooperativity. Other regions of Fos and Jun had smaller effects on both the orientation of heterodimer binding and on bending cooperativity. Consequently, both oriented heterodimer binding and cooperative DNA bending in the NFAT complex required the residues at the amino-terminal ends of the leucine zippers of both Fos and Jun.
DISCUSSION
The regulation of transcription initiation depends on interactions among multiple proteins that bind to separate recognition sites. Many eukaryotic transcription factors bind to palindromic regulatory elements as heteromeric complexes. Such complexes could in principle bind to their recognition sites in either orientation. Complexes that bind in opposite orientations are predicted to present different surfaces for interactions with transcription factors that bind to adjacent regulatory elements. Regulation of the orientation of transcription factor binding therefore provides a potential mechanism for control of transcription factor interactions and for modulation of transcriptional activity.
The NFAT1–Fos–Jun complex provides a model for understanding the influence of transcription factor interactions on the structural organization of transcription regulatory protein complexes. The results of phasing analysis demonstrated that cooperative DNA binding by Fos–Jun heterodimers and NFAT1 induced DNA bending toward the interaction interface. The results of fluorescence architectural analysis demonstrated that the interaction with NFAT1 controlled the orientation of heterodimer binding at the ARRE2 site. Thus, formation of the NFAT1—Fos–Jun—ARRE2 complex is associated with changes in both protein and DNA structure.
The structural changes identified in these studies are consistent with the recently solved x-ray crystal structure of the bZIP domains of Fos–Jun and the DNA binding region of NFAT1 bound to the ARRE2 site (24). The direction and magnitude of the cooperative DNA bend induced by Fos–Jun interaction with NFAT1 are consistent with the change in DNA structure required for the NFAT1—Fos–Jun interaction observed in the crystal. The overall DNA bend observed for the NFAT1—Fos–Jun—ARRE2 complex in solution is also similar in direction but approximately 50% greater in magnitude than the bend observed in the crystal. This DNA bend results from the combined effects of bending by Fos–Jun, bending by NFAT1, bending caused by the interaction of Fos–Jun with NFAT1, and the intrinsic bend at the ARRE2 site.
No DNA bending was observed in the NMR structure of a fragment of the NFAT2 (NFATc1) DNA binding region bound to a 12-bp oligonucleotide (25). The fragment of NFAT2 used in these studies contained a substitution at a key DNA contact residue and lacked other residues that contact DNA in the NFAT1—Fos–Jun—ARRE2 complex (24). It is therefore possible that the lack of DNA bending in the NMR structure is due to the short oligonucleotide binding site or the differences in DNA contacts.
The same amino acid residues in Fos and Jun determined both the orientation of heterodimer binding and the magnitude and direction of cooperative DNA bending in the NFAT1—Fos–Jun—ARRE2 complex. In the x-ray crystal structure, an extended interaction interface that involves a large number of residues over the entire length of the leucine zipper is observed (25). The results of our experiments identify a small subset of these residues as the principal determinants of the asymmetric interaction between Fos–Jun and NFAT1 (Fig. 4). The additional interactions may represent adventitious contacts that occur because of the proximity of the proteins, or they may be replaced by alternative contacts in heterodimers that bind in the opposite orientation. Our results are inconsistent with a previous model in which a single arginine in the spacer region of Jun was suggested to be both necessary and sufficient for interaction with NFAT2 (26). This arginine had no effect on the orientation of heterodimer binding or on bending cooperativity.
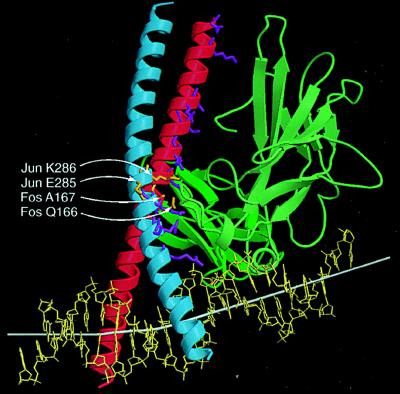
Figure 4.
Comparison of residues required for oriented heterodimer binding and cooperative DNA bending with contact residues in the x-ray crystal structure. The smoothed Cα trace of NFAT1 (green) and Fos–Jun (red-blue) are shown using ribbons to indicate α helices and arrows to indicate β sheets based on the x-ray crystal structure (24). The side chains of residues in Fos and Jun that are in close contact with NFAT1 in the x-ray crystal structure (24) are shown in stick representation. The subset of these residues that determined the orientation of heterodimer binding and cooperative DNA bending is shown in orange and indicated by white arrows. Other contact residues are shown in purple. The helix axis is shown by a white bar that is extended on both ends based on the three base pairs at the ends. The DNA helix axis was calculated using curves and the image was produced using molscript and raster3d.
The fluorescence architectural analysis provides a general method for determination of the structural organization of transcription regulatory protein complexes. Its advantages include the ability to compare fluorophores placed at different positions removed from interaction interface, the ability to quantitate structural differences between related complexes, and the ability to determine the conformation complexes in solution. Because our application of the fluorescence resonance energy transfer assay does not require measurement of exact distances between the fluorophores, it is less sensitive to the local environments and orientations of the fluorophores. This approach provides a powerful new tool for analysis of the structural organization of nucleoprotein complexes.
Transcription factor interactions are central to the cooperative regulation of gene expression (3, 4, 27). The NFAT1—Fos–Jun complex functions in the context of promoter regions containing binding sites for numerous other transcription regulatory proteins. Transcription factor binding at the interleukin-2 promoter in vivo is coordinately regulated in response to both activators and inhibitors of interleukin-2 transcription (28). It is therefore probable that transcription factor binding at the interleukin-2 promoter is stabilized through interactions among the proteins that bind to the individual regulatory elements as has been demonstrated for other promoter and enhancer regions (3, 4). These interactions are likely to require bending of the promoter region and the correct orientation of Fos–Jun heterodimer binding. Thus, the effects of interactions between Fos–Jun and NFAT1 on DNA bending and on the orientation of heterodimer binding may contribute to the appropriate structural organization of the promoter region and to the control of transcriptional activity.
Acknowledgments
We thank Mensur Dlakic for preparation of Fig. 4, Gerard Jenkins for assistance with plasmid construction, and Cassandra Wong for oligonucleotide synthesis.
ABBREVIATIONS
- NFAT1
nuclear factor of activated T cells 1
- FRET
fluorescence resonance energy transfer
- bZIP
basic region–leucine zipper
References
- 1.Jain J, McCaffrey P G, Miner Z, Kerppola T K, Lambert J N, Verdine G L, Curran T, Rao A. Nature. 1993;365:352–355. doi: 10.1038/365352a0. [DOI] [PubMed] [Google Scholar]
- 2.McCaffrey P G, et al. Science. 1993;262:750–754. doi: 10.1126/science.8235597. [DOI] [PubMed] [Google Scholar]
- 3.Giese K, Kingsley C, Kirshner J R, Grosschedl R. Genes Dev. 1995;9:995–1008. doi: 10.1101/gad.9.8.995. [DOI] [PubMed] [Google Scholar]
- 4.Kim T K, Maniatis T. Mol Cell. 1998;1:119–129. doi: 10.1016/s1097-2765(00)80013-1. [DOI] [PubMed] [Google Scholar]
- 5.Kerppola T K, Curran T. Cell. 1991;66:317–326. doi: 10.1016/0092-8674(91)90621-5. [DOI] [PubMed] [Google Scholar]
- 6.Kerppola T K, Curran T. Science. 1991;254:1210–1214. doi: 10.1126/science.1957173. [DOI] [PubMed] [Google Scholar]
- 7.Leonard D A, Rajaram N, Kerppola T K. Proc Natl Acad Sci USA. 1997;94:4913–4918. doi: 10.1073/pnas.94.10.4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kerppola T K. Biochemistry. 1997;36:10872–10884. doi: 10.1021/bi971119w. [DOI] [PubMed] [Google Scholar]
- 9.Rajaram N, Kerppola T K. EMBO J. 1997;16:2917–2925. doi: 10.1093/emboj/16.10.2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Strauss-Soukup J K, Maher L J., III Biochemistry. 1997;36:10026–10032. doi: 10.1021/bi970215u. [DOI] [PubMed] [Google Scholar]
- 11.Paolella D N, Liu Y, Fabian M A, Schepartz A. Biochemistry. 1997;36:10033–10038. doi: 10.1021/bi970515b. [DOI] [PubMed] [Google Scholar]
- 12.Strauss-Soukup J K, Maher L J., III Biochemistry. 1998;37:1060–1066. doi: 10.1021/bi972146p. [DOI] [PubMed] [Google Scholar]
- 13.Glover J N, Harrison S C. Nature. 1995;373:257–261. doi: 10.1038/373257a0. [DOI] [PubMed] [Google Scholar]
- 14.Sitlani A, Crothers D M. Proc Natl Acad Sci USA. 1998;95:1404–1409. doi: 10.1073/pnas.95.4.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kerppola T. Proc Natl Acad Sci USA. 1996;93:10117–10122. doi: 10.1073/pnas.93.19.10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kerppola T K, Curran T. EMBO J. 1997;16:2907–2916. doi: 10.1093/emboj/16.10.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leveillard T, Kassavetis G A, Geiduschek E P. J Biol Chem. 1991;367:5162–5168. [PubMed] [Google Scholar]
- 18.Oelgeschlager T, Chiang C M, Roeder R G. Nature. 1996;382:735–738. doi: 10.1038/382735a0. [DOI] [PubMed] [Google Scholar]
- 19.Forget D, Robert F, Grondin G, Burton Z F, Greenblatt J, Coulombe B. Proc Natl Acad Sci USA. 1997;94:7150–7155. doi: 10.1073/pnas.94.14.7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim T-K, Lagrange T, Wang Y-H, Griffith J D, Reinberg D, Ebright R H. Proc Natl Acad Sci USA. 1997;94:12268–12273. doi: 10.1073/pnas.94.23.12268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jain J, Burgeon E, Badalian T M, Hogan P G, Rao A. J Biol Chem. 1995;270:4138–4145. [PubMed] [Google Scholar]
- 22.Patel L R, Curran T, Kerppola T K. Proc Natl Acad Sci USA. 1994;91:7360–7364. doi: 10.1073/pnas.91.15.7360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen L, Oakley M G, Glover J N, Jain J, Dervan P B, Hogan P G, Rao A, Verdine G L. Curr Biol. 1995;5:882–889. doi: 10.1016/s0960-9822(95)00178-3. [DOI] [PubMed] [Google Scholar]
- 24.Chen L, Glover J N M, Hogan P G, Rao A, Harrison S C. Nature. 1998;392:42–48. doi: 10.1038/32100. [DOI] [PubMed] [Google Scholar]
- 25.Zhou P, Sun L J, Dotsch V, Wagner G, Verdine G L. Cell. 1998;92:687–696. doi: 10.1016/s0092-8674(00)81136-8. [DOI] [PubMed] [Google Scholar]
- 26.Peterson B R, Sun L J, Verdine G L. Proc Natl Acad Sci USA. 1996;93:13671–13676. doi: 10.1073/pnas.93.24.13671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robertson L M, Kerppola T K, Vendrell M, Luk D, Smeyne R J, Bocchiaro C, Morgan J I, Curran T. Neuron. 1995;14:241–252. doi: 10.1016/0896-6273(95)90282-1. [DOI] [PubMed] [Google Scholar]
- 28.Garrity P A, Chen D, Rothenberg E V, Wold B J. Mol Cell Biol. 1994;14:2159–2169. doi: 10.1128/mcb.14.3.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]