Abstract
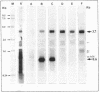
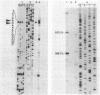
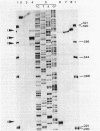
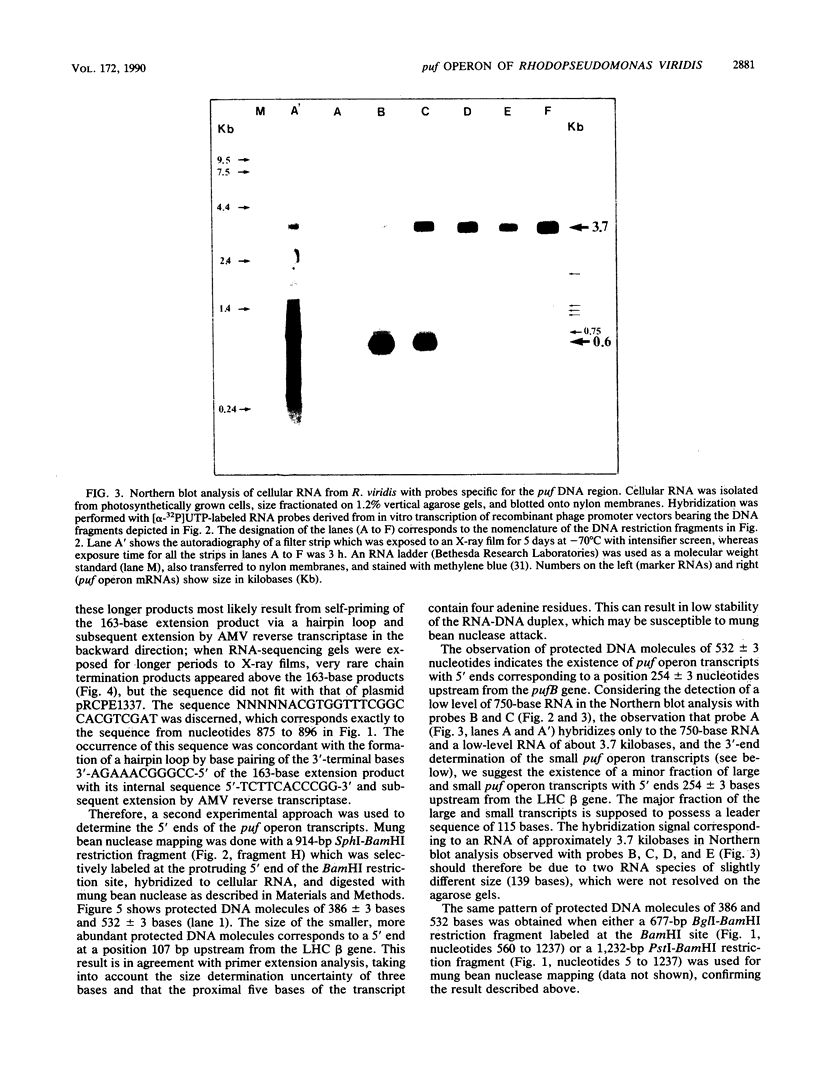
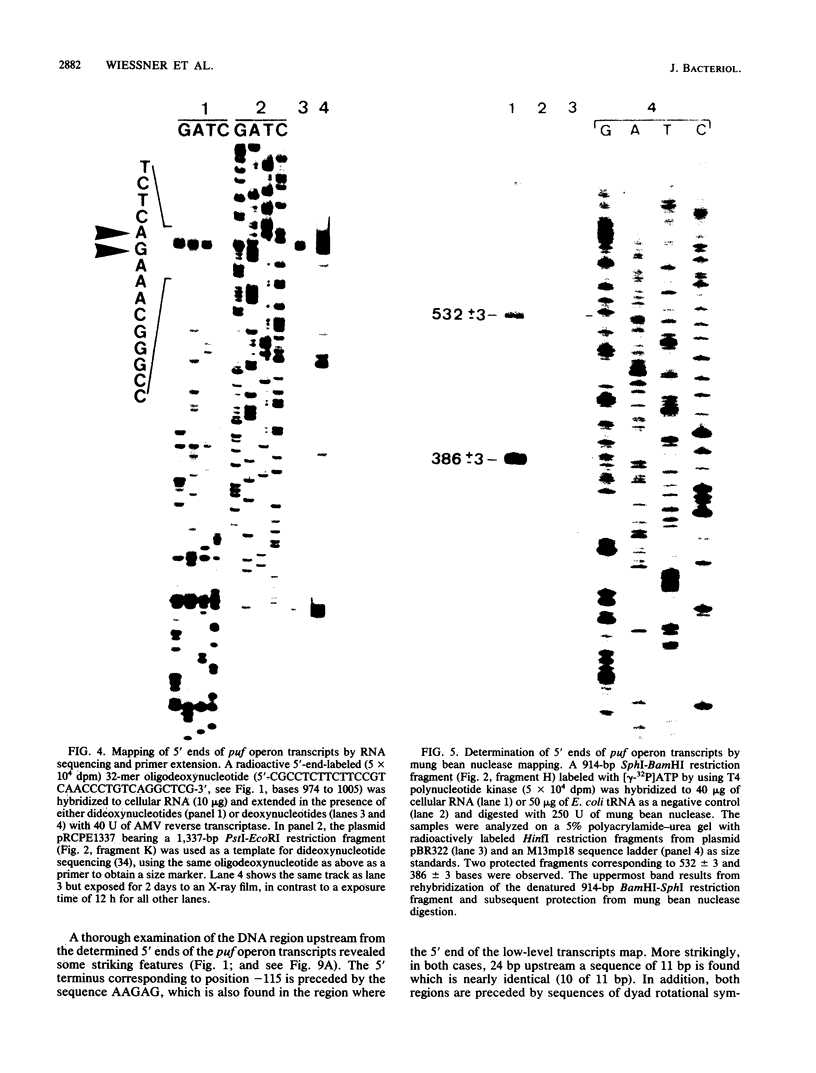
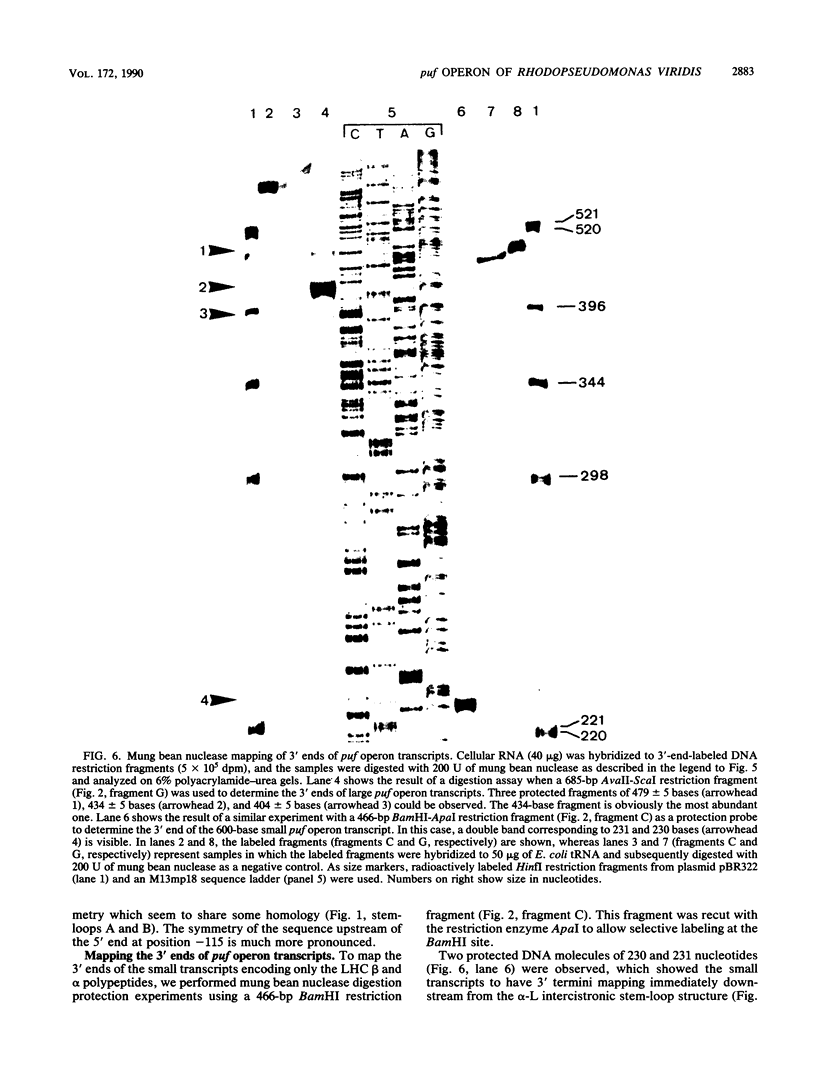
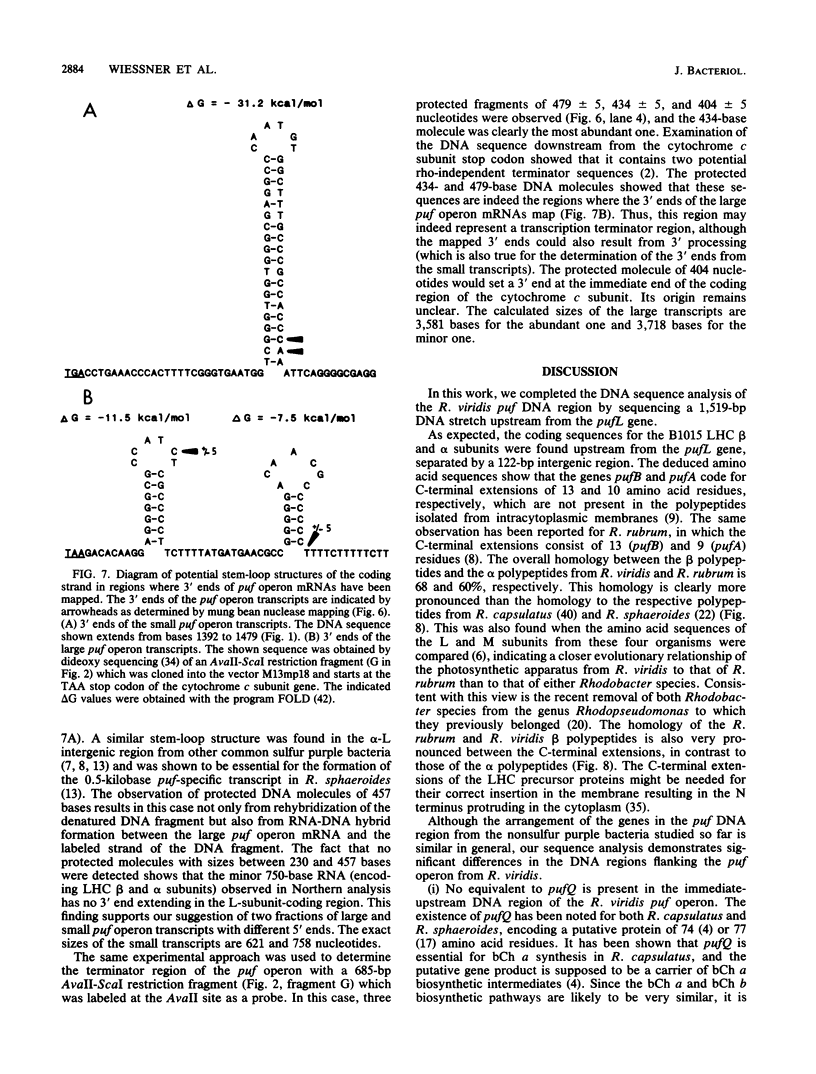
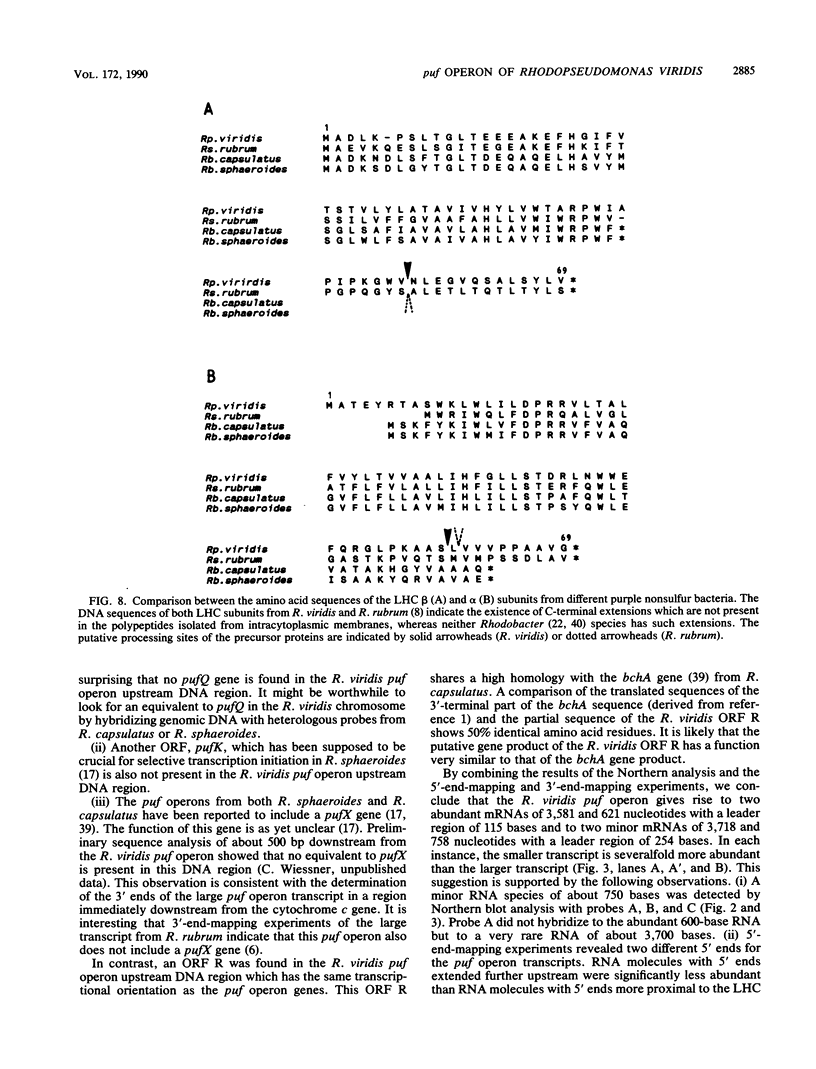
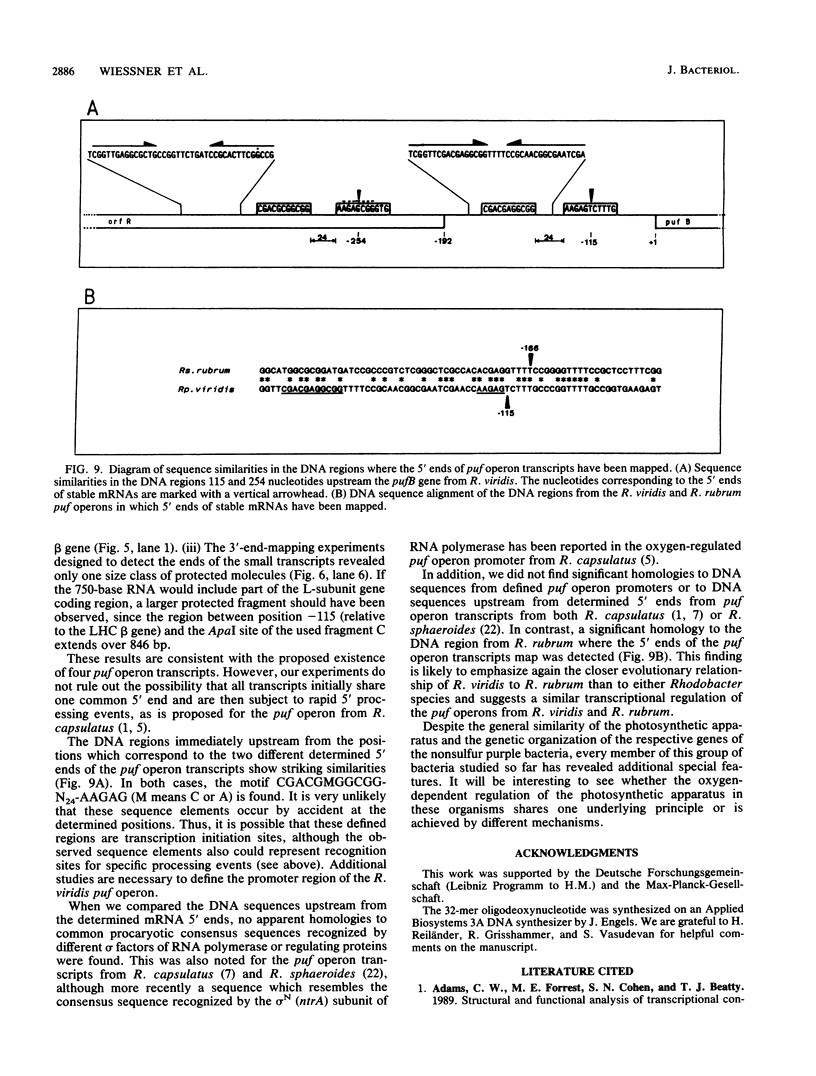
The genes encoding the beta and alpha subunits of the B1015 light-harvesting complex (LHC) and the L, M, and cytochrome c subunits of the photosynthetic reaction center from Rhodopseudomonas viridis are organized in an operon, in analogy to other nonsulfur purple bacteria, named the puf operon. In photoheterotrophically grown cells, two abundant puf operon mRNA species of 3,581 and 621 bases were present. The large transcript encoded the LHC beta, LHC alpha, and reaction center L, M, and cytochrome c polypeptides, whereas the small transcript only coded for the LHC beta and alpha polypeptides. Both transcripts share a common 5' end which is located 115 bases upstream from the initiation codon of the LHC beta gene. Two additional low-level transcripts of 3,718 and 758 bases with 5' ends 254 +/- 3 bases upstream from the LHC beta gene were detected. Analysis of the DNA sequence preceding the different 5' ends revealed DNA elements of striking homology. The 3' ends of the small transcripts were mapped within the alpha-L intercistronic DNA region downstream from a sequence capable of forming a very stable stem-loop when transcribed into RNA. The 3' termini of the large transcripts are located immediately downstream from the region coding the cytochrome c subunit in two areas resembling rho-independent transcription terminators. No open reading frames corresponding to pufQ and pufX from Rhodobacter capsulatus and Rhodobacter sphaeroides were present in the flanking DNA regions of the puf operon. In contrast, an open reading frame ending 191 base pairs upstream from the LHC beta gene showed 50% homology at the amino acid level to the available sequence of the bchA gene from R. capsulatus. The genes coding for the B1015 LHC subunits had C-terminal extensions of 13 (beta) and 10 (alpha) amino acids which were not present in the proteins isolated from intracytoplasmic membranes.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams C. W., Forrest M. E., Cohen S. N., Beatty J. T. Structural and functional analysis of transcriptional control of the Rhodobacter capsulatus puf operon. J Bacteriol. 1989 Jan;171(1):473–482. doi: 10.1128/jb.171.1.473-482.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adhya S., Gottesman M. Control of transcription termination. Annu Rev Biochem. 1978;47:967–996. doi: 10.1146/annurev.bi.47.070178.004535. [DOI] [PubMed] [Google Scholar]
- Bauer C. E., Marrs B. L. Rhodobacter capsulatus puf operon encodes a regulatory protein (PufQ) for bacteriochlorophyll biosynthesis. Proc Natl Acad Sci U S A. 1988 Oct;85(19):7074–7078. doi: 10.1073/pnas.85.19.7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer C. E., Young D. A., Marrs B. L. Analysis of the Rhodobacter capsulatus puf operon. Location of the oxygen-regulated promoter region and the identification of an additional puf-encoded gene. J Biol Chem. 1988 Apr 5;263(10):4820–4827. [PubMed] [Google Scholar]
- Belasco J. G., Beatty J. T., Adams C. W., von Gabain A., Cohen S. N. Differential expression of photosynthesis genes in R. capsulata results from segmental differences in stability within the polycistronic rxcA transcript. Cell. 1985 Jan;40(1):171–181. doi: 10.1016/0092-8674(85)90320-4. [DOI] [PubMed] [Google Scholar]
- Brunisholz R. A., Jay F., Suter F., Zuber H. The light-harvesting polypeptides of Rhodopseudomonas viridis. The complete amino-acid sequences of B1015-alpha, B1015-beta and B1015-gamma. Biol Chem Hoppe Seyler. 1985 Jan;366(1):87–98. doi: 10.1515/bchm3.1985.366.1.87. [DOI] [PubMed] [Google Scholar]
- Bélanger G., Gingras G. Structure and expression of the puf operon messenger RNA in rhodospirillum rubrum. J Biol Chem. 1988 Jun 5;263(16):7639–7645. [PubMed] [Google Scholar]
- Bérard J., Bélanger G., Corriveau P., Gingras G. Molecular cloning and sequence of the B880 holochrome gene from Rhodospirillum rubrum. J Biol Chem. 1986 Jan 5;261(1):82–87. [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Davis J., Donohue T. J., Kaplan S. Construction, characterization, and complementation of a Puf- mutant of Rhodobacter sphaeroides. J Bacteriol. 1988 Jan;170(1):320–329. doi: 10.1128/jb.170.1.320-329.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeHoff B. S., Lee J. K., Donohue T. J., Gumport R. I., Kaplan S. In vivo analysis of puf operon expression in Rhodobacter sphaeroides after deletion of a putative intercistronic transcription terminator. J Bacteriol. 1988 Oct;170(10):4681–4692. doi: 10.1128/jb.170.10.4681-4692.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deisenhofer J., Epp O., Miki K., Huber R., Michel H. X-ray structure analysis of a membrane protein complex. Electron density map at 3 A resolution and a model of the chromophores of the photosynthetic reaction center from Rhodopseudomonas viridis. J Mol Biol. 1984 Dec 5;180(2):385–398. doi: 10.1016/s0022-2836(84)80011-x. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drews G., Giesbrecht P. Rhodopseudomonas viridis, nov. spec., ein neu isoliertes, obligat phototrophes Bakterium. Arch Mikrobiol. 1966 Mar 31;53(3):255–262. [PubMed] [Google Scholar]
- Kiley P. J., Donohue T. J., Havelka W. A., Kaplan S. DNA sequence and in vitro expression of the B875 light-harvesting polypeptides of Rhodobacter sphaeroides. J Bacteriol. 1987 Feb;169(2):742–750. doi: 10.1128/jb.169.2.742-750.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiley P. J., Kaplan S. Molecular genetics of photosynthetic membrane biosynthesis in Rhodobacter sphaeroides. Microbiol Rev. 1988 Mar;52(1):50–69. doi: 10.1128/mr.52.1.50-69.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang F. S., Oesterhelt D. Microaerophilic growth and induction of the photosynthetic reaction center in Rhodopseudomonas viridis. J Bacteriol. 1989 May;171(5):2827–2834. doi: 10.1128/jb.171.5.2827-2834.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel H. Three-dimensional crystals of a membrane protein complex. The photosynthetic reaction centre from Rhodopseudomonas viridis. J Mol Biol. 1982 Jul 5;158(3):567–572. doi: 10.1016/0022-2836(82)90216-9. [DOI] [PubMed] [Google Scholar]
- Michel H., Weyer K. A., Gruenberg H., Dunger I., Oesterhelt D., Lottspeich F. The 'light' and 'medium' subunits of the photosynthetic reaction centre from Rhodopseudomonas viridis: isolation of the genes, nucleotide and amino acid sequence. EMBO J. 1986 Jun;5(6):1149–1158. doi: 10.1002/j.1460-2075.1986.tb04340.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel H., Weyer K. A., Gruenberg H., Lottspeich F. The ;heavy' subunit of the photosynthetic reaction centre from Rhodopseudomonas viridis: isolation of the gene, nucleotide and amino acid sequence. EMBO J. 1985 Jul;4(7):1667–1672. doi: 10.1002/j.1460-2075.1985.tb03835.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince R. C., Leigh J. S., Jr, Dutton P. L. Thermodynamic properties of the reaction center of Rhodopseudomonas viridis. In vivo measurement of the reaction center bacteriochlorophyll-primary acceptor intermediary electron carrier. Biochim Biophys Acta. 1976 Sep 13;440(3):622–636. doi: 10.1016/0005-2728(76)90047-5. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornber J. P., Cogdell R. J., Seftor R. E., Webster G. D. Further studies on the composition and spectral properties of the photochemical reaction centers of bacteriochlorophyll b-containing bacteria. Biochim Biophys Acta. 1980 Nov 5;593(1):60–75. doi: 10.1016/0005-2728(80)90008-0. [DOI] [PubMed] [Google Scholar]
- Weyer K. A., Lottspeich F., Gruenberg H., Lang F., Oesterhelt D., Michel H. Amino acid sequence of the cytochrome subunit of the photosynthetic reaction centre from the purple bacterium Rhodopseudomonas viridis. EMBO J. 1987 Aug;6(8):2197–2202. doi: 10.1002/j.1460-2075.1987.tb02490.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young D. A., Bauer C. E., Williams J. C., Marrs B. L. Genetic evidence for superoperonal organization of genes for photosynthetic pigments and pigment-binding proteins in Rhodobacter capsulatus. Mol Gen Genet. 1989 Jul;218(1):1–12. doi: 10.1007/BF00330558. [DOI] [PubMed] [Google Scholar]
- Youvan D. C., Bylina E. J., Alberti M., Begusch H., Hearst J. E. Nucleotide and deduced polypeptide sequences of the photosynthetic reaction-center, B870 antenna, and flanking polypeptides from R. capsulata. Cell. 1984 Jul;37(3):949–957. doi: 10.1016/0092-8674(84)90429-x. [DOI] [PubMed] [Google Scholar]
- Zhu Y. S., Hearst J. E. Regulation of expression of genes for light-harvesting antenna proteins LH-I and LH-II; reaction center polypeptides RC-L, RC-M, and RC-H; and enzymes of bacteriochlorophyll and carotenoid biosynthesis in Rhodobacter capsulatus by light and oxygen. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7613–7617. doi: 10.1073/pnas.83.20.7613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]