Abstract
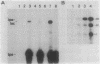
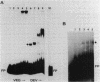
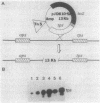
A segment of DNA located between 131 and 311 base pairs (bp) upstream from the transcriptional start of the Myxococcus xanthus ops gene (-131 to -311) was shown to function as an upstream activation site (UAS) for developmentally regulated transcription from the tps gene promoter region. The activation of early developmental transcription by the ops UAS was independent of orientation and could be increased by the addition of a second copy of the UAS. The ops UAS segment continued to function when placed 1.5 kbp upstream from the transcription initiation site. DNA from the tps promoter region was required for transcriptional activation by the ops UAS, and a specific requirement for the sequence of tps DNA between -34 and -66 was demonstrated. Several specific ops UAS DNA-protein complexes were observed after incubation of this DNA segment with an extract of early developmental M. xanthus cells. Extracts of vegetative cells contained much less ops UAS-specific DNA-binding activity. When the distance between the tps and ops genes was increased from 2 to 15 kbp by insertion of a transduced segment of DNA, the amount of developmentally induced tps RNA was found to be about one-third that found in wild-type M. xanthus. Our observations suggest that the regulatory region of the ops gene functions not only to control ops gene expression but also to increase early developmental expression of the tps gene located about 2 kbp downstream on the M. xanthus chromosome.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Atchison M. L. Enhancers: mechanisms of action and cell specificity. Annu Rev Cell Biol. 1988;4:127–153. doi: 10.1146/annurev.cb.04.110188.001015. [DOI] [PubMed] [Google Scholar]
- Campos J. M., Geisselsoder J., Zusman D. R. Isolation of bacteriophage MX4, a generalized transducing phage for Myxococcus xanthus. J Mol Biol. 1978 Feb 25;119(2):167–178. doi: 10.1016/0022-2836(78)90431-x. [DOI] [PubMed] [Google Scholar]
- DWORKIN M., GIBSON S. M. A SYSTEM FOR STUDYING MICROBIAL MORPHOGENESIS: RAPID FORMATION OF MICROCYSTS IN MYXOCOCCUS XANTHUS. Science. 1964 Oct 9;146(3641):243–244. doi: 10.1126/science.146.3641.243. [DOI] [PubMed] [Google Scholar]
- Downard J. S. Identification of the RNA products of the ops gene of Myxococcus xanthus and mapping of ops and tps RNAs. J Bacteriol. 1987 Apr;169(4):1522–1528. doi: 10.1128/jb.169.4.1522-1528.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downard J. S., Kim S. H., Kil K. S. Localization of the cis-acting regulatory DNA sequences of the Myxococcus xanthus tps and ops genes. J Bacteriol. 1988 Oct;170(10):4931–4938. doi: 10.1128/jb.170.10.4931-4938.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downard J. S., Kupfer D., Zusman D. R. Gene expression during development of Myxococcus xanthus. Analysis of the genes for protein S. J Mol Biol. 1984 Jun 5;175(4):469–492. doi: 10.1016/0022-2836(84)90180-3. [DOI] [PubMed] [Google Scholar]
- Downard J. S. Tn5-mediated transposition of plasmid DNA after transduction to Myxococcus xanthus. J Bacteriol. 1988 Oct;170(10):4939–4941. doi: 10.1128/jb.170.10.4939-4941.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downard J. S., Zusman D. R. Differential expression of protein S genes during Myxococcus xanthus development. J Bacteriol. 1985 Mar;161(3):1146–1155. doi: 10.1128/jb.161.3.1146-1155.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried M., Crothers D. M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981 Dec 11;9(23):6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner M. M., Revzin A. A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the Escherichia coli lactose operon regulatory system. Nucleic Acids Res. 1981 Jul 10;9(13):3047–3060. doi: 10.1093/nar/9.13.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill R. E., Cull M. G. Control of developmental gene expression by cell-to-cell interactions in Myxococcus xanthus. J Bacteriol. 1986 Oct;168(1):341–347. doi: 10.1128/jb.168.1.341-347.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross C., Engbaek F., Flammang T., Burgess R. Rapid micromethod for the purification of Escherichia coli ribonucleic acid polymerase and the preparation of bacterial extracts active in ribonucleic acid synthesis. J Bacteriol. 1976 Oct;128(1):382–389. doi: 10.1128/jb.128.1.382-389.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen D. C., Bretscher A. P., Kaiser D. Synergism between morphogenetic mutants of Myxococcus xanthus. Dev Biol. 1978 Jun;64(2):284–296. doi: 10.1016/0012-1606(78)90079-9. [DOI] [PubMed] [Google Scholar]
- Inouye M., Inouye S., Zusman D. R. Biosynthesis and self-assembly of protein S, a development-specific protein of Myxococcus xanthus. Proc Natl Acad Sci U S A. 1979 Jan;76(1):209–213. doi: 10.1073/pnas.76.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye S., Franceschini T., Inouye M. Structural similarities between the development-specific protein S from a gram-negative bacterium, Myxococcus xanthus, and calmodulin. Proc Natl Acad Sci U S A. 1983 Nov;80(22):6829–6833. doi: 10.1073/pnas.80.22.6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye S. Identification of a development-specific promoter of Myxococcus xanthus. J Mol Biol. 1984 Mar 25;174(1):113–120. doi: 10.1016/0022-2836(84)90367-x. [DOI] [PubMed] [Google Scholar]
- Inouye S., Ike Y., Inouye M. Tandem repeat of the genes for protein S, a development-specific protein of Myxococcus xanthus. J Biol Chem. 1983 Jan 10;258(1):38–40. [PubMed] [Google Scholar]
- Kaiser D. Control of multicellular development: Dictyostelium and Myxococcus. Annu Rev Genet. 1986;20:539–566. doi: 10.1146/annurev.ge.20.120186.002543. [DOI] [PubMed] [Google Scholar]
- Kaiser D., Manoil C., Dworkin M. Myxobacteria: cell interactions, genetics, and development. Annu Rev Microbiol. 1979;33:595–639. doi: 10.1146/annurev.mi.33.100179.003115. [DOI] [PubMed] [Google Scholar]
- Kroos L., Kaiser D. Expression of many developmentally regulated genes in Myxococcus depends on a sequence of cell interactions. Genes Dev. 1987 Oct;1(8):840–854. doi: 10.1101/gad.1.8.840. [DOI] [PubMed] [Google Scholar]
- Kuspa A., Kroos L., Kaiser D. Intercellular signaling is required for developmental gene expression in Myxococcus xanthus. Dev Biol. 1986 Sep;117(1):267–276. doi: 10.1016/0012-1606(86)90369-6. [DOI] [PubMed] [Google Scholar]
- Kustu S., Santero E., Keener J., Popham D., Weiss D. Expression of sigma 54 (ntrA)-dependent genes is probably united by a common mechanism. Microbiol Rev. 1989 Sep;53(3):367–376. doi: 10.1128/mr.53.3.367-376.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRossa R., Kuner J., Hagen D., Manoil C., Kaiser D. Developmental cell interactions of Myxococcus xanthus: analysis of mutants. J Bacteriol. 1983 Mar;153(3):1394–1404. doi: 10.1128/jb.153.3.1394-1404.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson D. R., Zusman D. R. Transport and localization of protein S, a spore coat protein, during fruiting body formation by Myxococcus xanthus. J Bacteriol. 1983 May;154(2):547–553. doi: 10.1128/jb.154.2.547-553.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninfa A. J., Magasanik B. Covalent modification of the glnG product, NRI, by the glnL product, NRII, regulates the transcription of the glnALG operon in Escherichia coli. Proc Natl Acad Sci U S A. 1986 Aug;83(16):5909–5913. doi: 10.1073/pnas.83.16.5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor K. A., Zusman D. R. Coliphage P1-mediated transduction of cloned DNA from Escherichia coli to Myxococcus xanthus: use for complementation and recombinational analyses. J Bacteriol. 1983 Jul;155(1):317–329. doi: 10.1128/jb.155.1.317-329.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popham D. L., Szeto D., Keener J., Kustu S. Function of a bacterial activator protein that binds to transcriptional enhancers. Science. 1989 Feb 3;243(4891):629–635. doi: 10.1126/science.2563595. [DOI] [PubMed] [Google Scholar]
- Struhl K. Molecular mechanisms of transcriptional regulation in yeast. Annu Rev Biochem. 1989;58:1051–1077. doi: 10.1146/annurev.bi.58.070189.005155. [DOI] [PubMed] [Google Scholar]
- Teintze M., Thomas R., Furuichi T., Inouye M., Inouye S. Two homologous genes coding for spore-specific proteins are expressed at different times during development of Myxococcus xanthus. J Bacteriol. 1985 Jul;163(1):121–125. doi: 10.1128/jb.163.1.121-125.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]