Abstract
Hypoxia induces a group of physiologically important genes such as erythropoietin and vascular endothelial growth factor. These genes are transcriptionally up-regulated by hypoxia-inducible factor 1 (HIF-1), a global regulator that belongs to the basic helix-loop-helix PAS family. Although HIF-1 is a heterodimer composed of α and β subunits, its activity is primarily determined by hypoxia-induced stabilization of HIF-1α, which is otherwise rapidly degraded in oxygenated cells. We report the identification of an oxygen-dependent degradation (ODD) domain within HIF-1α that controls its degradation by the ubiquitin-proteasome pathway. The ODD domain consists of ≈200 amino acid residues, located in the central region of HIF-1α. Because portions of the domain independently confer degradation of HIF-1α, deletion of this entire region is required to give rise to a stable HIF-1α, capable of heterodimerization, DNA-binding, and transactivation in the absence of hypoxic signaling. Conversely, the ODD domain alone confers oxygen-dependent instability when fused to a stable protein, Gal4. Hence, the ODD domain plays a pivotal role for regulating HIF-1 activity and thereby may provide a means of controlling gene expression by changes in oxygen tension.
The viability and function of both individual cells and the organism as a whole depend on the modulation of a variety of genes in response to changes in oxygen tension. In the last decade, there has been substantial progress in understanding the molecular basis of oxygen sensing and transcriptional regulation of physiologically relevant genes including those encoding erythropoietin, vascular endothelial growth factor, tyrosine hydroxylase, inducible nitric oxide synthase and glycolytic enzymes (for reviews see refs. 1 and 2). Induction of these genes confer multiple responses to maintain oxygen homeostasis. Remarkably, at the transcriptional level these diverse genes are all under the control of a crucial transcription factor: hypoxia-inducible factor 1 (HIF-1) (3, 4).
HIF-1 is an αβ heterodimer, both subunits of which belong to the rapidly growing PAS family of basic helix-loop-helix (bHLH) transcription factors (5). The β subunit is identical to the aryl hydrocarbon nuclear translocator (ARNT) that also serves as a heterodimeric partner with the aryl hydrocarbon receptor (6). In contrast, it appears that the α subunit’s sole but critical function is mediation of the response to hypoxia. Like most members of this family, near the N terminus both subunits have a bHLH domain that is necessary for DNA binding (7, 8). Downstream PAS domains are required for dimerization (7–10) and target gene specificity (11). The C-terminal portion varies greatly among members of the PAS family. Although ARNT contains a transactivation domain near the C terminus (12, 13), it does not appear to be necessary for HIF-1 transactivation (14). Recently, two transactivation domains on HIF-1α have been identified, one at the C terminus and the other in the middle of the polypeptide (15–17).
Despite these findings, there is very little understanding of the steps underlying the activation of HIF-1 by hypoxia. Phosphorylation may be critical in the signaling process (4, 18), but there is no evidence thus far that HIF-1 phosphorylation is hypoxia-dependent. Binding of HSP90 with HIF-1α has been demonstrated in vitro (19), and yet, the biological significance of this interaction remains to be determined. Recently, ARNT has been shown to protect proteolysis of HIF-1α in vitro (20). Moreover, several groups reported that the activity of HIF-1α transactivation domains in Gal4 fusions is hypoxia inducible (15–17). Although all of these observations are important, taken together, they do not suggest a coherent model for the activation pathway of HIF-1 in response to hypoxia.
Altering oxygen tension has no significant effect on steady-state levels of either HIF-1α mRNA or ARNT mRNA (14, 19–21). Moreover, as was reported (21), at the protein level ARNT is constitutively expressed and not significantly affected by oxygen. In contrast, HIF-1α protein is remarkably unstable in cells exposed to oxygen, whereas hypoxia induces a striking increase in the abundance of HIF-1α protein. These observations suggested that accumulation of HIF-1α is a prerequisite to the activation of HIF-1 and led to a hypothesis that the activation of HIF-1 depends primarily on hypoxia-induced stabilization of HIF-1α, which is otherwise rapidly degraded in oxygenated cells (21). In this paper, we present evidence that unstable HIF-1α is controlled by an oxygen-dependent degradation (ODD) domain within HIF-1α and the degradation is catabolized through the ubiquitin-proteasome pathway. Removal of the entire ODD domain renders HIF-1α stable even in oxygenated cells, resulting in autonomous heterodimerization, DNA binding, and transactivation independent of hypoxia signaling.
MATERIALS AND METHODS
Cell Cultures and Reagents.
Hep3B cells were cultured in alpha-modified Eagle’s medium (GIBCO/BRL), and 293 cells in Dulbecco’s modified Eagle’s medium (GIBCO/BRL), supplemented with 10% fetal bovine serum (HyClone). Normoxia was defined as 95% air and 5% CO2 whereas hypoxia was defined as 1% oxygen, 94% N2, and 5% CO2. E64 [trans-epoxysuccinyl-l-leucylamido-(4-guanidino) butane], N-acetyl-l-leucinyl-l-leucinyl-methioninal, N-acetyl-l-leucinyl-l-leucinyl-l-l-norleucinal, and N-carbobenzoxyl-l-leucinyl-l-leucinyl-l-norvalinal (Cbz-LLL) were purchased from Sigma and dissolved in dimethyl sulfoxide. Cycloheximide and desferrioxamine were also from Sigma.
Plasmids.
The Gal4 DNA binding domain (1–147) in pCMXGal4(N) (22) was fused in frame to HIF-1α to create serial Gal4-HIF-1α fusions. Various portions of HIF-1α were PCR-amplified and cloned into the vector. HIF-1α(aa 530–814) fragment was purified from digested pHIF-1α (21) and cloned into the vector.
A synthesized hemagglutinin (HA) tag with NcoI site was inserted into pBS-HIF-1α and pBS-HIF-1α/SX, respectively (21). The HA-tagged HIF-1α was then transferred into pcDNA3 (Invitrogen), resulting in p(HA)HIF-1α and p(HA)HIF-1α(653). p(HA)HIF-1α(521) was made by PCR-amplification of HIF-1α(aa 335–521) with the creation of 3′ XbaI site, digestion with EcoRI and XbaI, and insertion into the digested p(HA)HIF-1α. The rest of C-terminal deletions were made by PCR-amplification to the desired codons. Internal deletions were essentially made by using the standard PCR amplification with two sets of primers. The resultant products were inserted into pcDNA3. All the constructs were confirmed by DNA sequencing.
pARNT was constructed by inserting the ARNT cDNA clone (provided by Oliver Hankinson, University of California, Los Angeles) into pcDNA3. The HA-tagged ubiquitin expression vector (pMT123) was provided by Dirk Bohmann (European Molecular Biology Laboratory, Heidelberg, Germany).
Transient Transfection and Preparation of Whole Cell Extracts.
293 cells were transfected in suspension using the calcium phosphate method as described elsewhere (21). A typical transfection was performed by using 4 μg pHIF-1α and 0.25 μg plasmid cytomegalovirus-β-galactosidase (β-gal). For hypoxia treatment, cells were incubated in a hypoxic chamber (Tapei/Espec, Osaka, Japan) for 4–6 hr before harvest. In some experiments, cells were pretreated with 60 μg/ml of cycloheximide for 15 min before exposure to hypoxia. Whole cell extracts were prepared essentially as described (21). Hep3B cells were transfected typically with 1 μg of pEpoEluc (21) and plasmid cytomegalovirus-β-gal, and 2 μg of p(HA)HIF-1α and pARNT for each 60 mm dish. Samples for hypoxia treatment were incubated overnight in the hypoxic chamber. Cells were lysed and assayed for luciferase activity as described elsewhere (21). β-Gal assays were performed for normalization of transfection efficiency.
Electrophoretic Mobility Shift Assay.
HIF-1 DNA binding reaction was performed as described (21). For Gal4 DNA binding reaction, 10–15 μg of cell extracts (normalized with β-gal assay) were mixed with 1 μg poly dI-dC in DNA binding for 5 min, followed by incubation for 10 min with 20 fmol 32P-labeled oligonucleotides containing Gal4 binding site.
Western Blot Analysis.
Western blot analysis was performed by using 20–30 μg of whole cell extracts normalized by β-gal assays as described before (21). Gal4-HIF-1α fusions were probed with 1:2,000 dilution of polyclonal anti-Gal4 antibody (Santa Cruz Biotechnology), and HA-tagged ubiquitin and HA-tagged HIF-1α were detected by using monoclonal anti-HA 12CA5 (Boehringer Mannheim). The antigen-antibody complexes were visualized by enhanced chemiluminescence (Amersham).
Metabolic Labeling, Pulse-Chase and Immunoprecipitation.
Metabolic radiolabeling of cells with [35S]methionine, pulse-chase and immunoprecipitation were performed as described (23). Cells were incubated with 100 mCi (1 Ci = 37 GBq) of [35S]methionine and [35S]cysteine (Tran35S-label, ICN) under either 21% O2 or 1% O2 for 4 hr. Samples were harvested and lysed in 1 ml of SDS-lysis buffer (23) containing the protease inhibitor mixture tablet Complete (Boehringer Mannheim). The intensity of the band was quantified by using PhosphorImager (Molecular Dynamics).
RESULTS
HIF-1α Is Unstable and Degraded Through the Ubiquitin- Proteasome Pathway.
HIF-1 DNA binding activity can be induced by exposing cells to 1% O2 (hypoxia), but upon switching to 21% O2 (normoxia) the binding activity quickly disappears owing to the rapid decay of HIF-1α subunit (21). Pulse-chase metabolic labeling with [35S]methionine confirmed that hypoxia-induced HIF-1α has a short half-life (<5 min) under normoxia (data not shown).
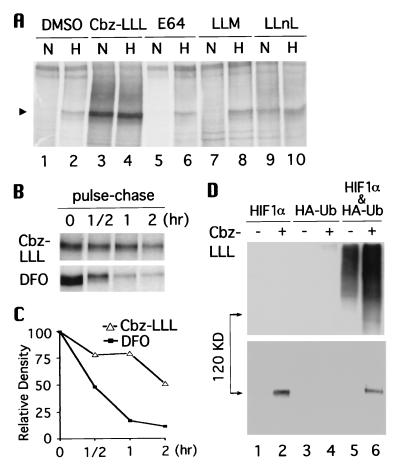
To elucidate the mechanism responsible for HIF-1α degradation, we employed peptide aldehydes that specifically inhibit the lysosomal proteases, calpains, and proteasome pathways (24). Hep3B cells were pretreated for 16 hr with inhibitors that target proteasomes (Cbz-LLL), lysosomal proteases [trans-epoxysuccinyl-l-leucylamido-(4-guanidino) butane] (E64), or calpains (N-acetyl-l-leucinyl-l-leucinyl-methioninal) (LLM), or with a less potent proteasome inhibitor (N-acetyl-l-leucinyl-l-leucinyl-l-norleucinal) (LLnL), and were either maintained under normoxia or incubated under hypoxia for an additional 4 hr. As analyzed by immunoprecipitation (Fig. 1A), HIF-1α was stabilized significantly under normoxia only by the proteasome inhibitor Cbz-LLL and N-acetyl-l-leucinyl-l-leucinyl-l-norleucinal (lanes 3 and 9), indicating that degradation of HIF-1α is mediated specifically via the proteasome pathway. A time-course experiment showed that after only 2-hr treatment with Cbz-LLL, HIF-1α became detectable under normoxia (data not shown). Furthermore, pulse-chase experiments (Fig. 1B) revealed that under normoxia the half-life of HIF-1α was prolonged to 2 hr after Cbz-LLL treatment, while desferrioxamine, an iron chelator known to mimic hypoxia (25), extended the half-life to only ≈30 min (Fig. 1C).
Figure 1.
Degradation of HIF-1α is via the ubiquitin-proteasome pathway. (A) Hep3B cells were pretreated with dimethyl sulfoxide, 10 μM Cbz-LLL, and 50 μM trans-epoxysuccinyl-l-leucylamido-(4-guanidino) butane, N-acetyl-l-leucinyl-l-leucinyl-methioninal, and a N-acetyl-l-leucinyl-l-leucinyl-l-norleucinal, respectively, for 16 hr, and radiolabeled with [35S]methionine in the presence of these reversible (39) inhibitors under normoxia (N) or hypoxia (H) for another 4 hr. Arrow heads indicate HIF-1α. B. Hep3B cells were incubated under normoxia with 10 μM Cbz-LLL (Upper) or 130 μM desferrioxamine (DFO) for 4 hr in the presence of [35S]methionine, and then chased for 0.5, 1, and 2 hr. Cell lysates were immunoprecipitated with anti-HIF-1α antibodies. The intensity of HIF-1α signals in B was quantified and plotted in C. (D) 293 cells were transfected with HIF-1α expression vector, HA-tagged ubiquitin vector, or both, and were treated with Cbz-LLL (+) or dimethyl sulfoxide (−) for 16 hr before harvest. Expressed HIF-1α was immunoprecipitated with anti-HIF-1α, determined by Western blot analysis with anti-HA antibody (Upper), and reprobed with anti-HIF-1α antibodies (Lower).
To determine if ubiquitination is involved in the degradation, 293 cells were transfected with expression vectors for HIF-1α or HA-tagged ubiquitin, or both (Fig. 1D). To maximize the levels of ubiquitin-conjugated HIF-1α intermediates, Cbz-LLL was used to block the proteasome pathway. HIF-1α was first immunoprecipitated by antibodies that specifically bind to HIF-1α and then probed with anti-HA antibody in Western blot. A smear of heavily ubiquitin-conjugated HIF-1α was detected only after both expression vectors were transfected, particularly in the presence of Cbz-LLL (Upper, lanes 5 and 6). The same blot was reprobed with the anti-HIF-1α antibodies to confirm equivalent levels of HIF-1α in the presence or absence of HA-tagged ubiquitin (Lower). Together, these results demonstrate that degradation of HIF-1α is mediated through the ubiquitin-dependent proteasome pathway.
Degradation of HIF-1α Is Controlled by an Oxygen-Dependent Degradation Domain.
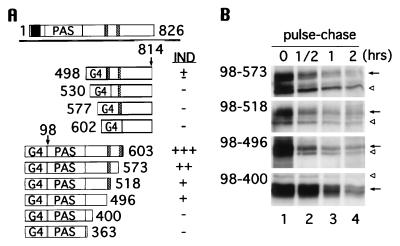
In search of regions within HIF-1α that regulate its degradation by the ubiquitin-proteasome pathway, we made use of Gal4 fusion expression vectors in which the Gal4 DNA binding domain was linked in frame with portions of HIF-1α (Fig. 2A). These constructs were transfected into 293 cells and whole cell extracts were analyzed by electrophoretic mobility shift of a radiolabeled Gal4 oligonucleotide as well as by Western blotting with anti-Gal4 antibodies. We began by focusing on the central region containing the two proline, glutamic acid, serine, threonine (PEST)-like sequences (5), which are often critical in protein degradation (26, 27).
Figure 2.
Identification of the regions responsible for oxygen-induced degradation by means of Gal4 fusion proteins. (A) The full-length HIF-1α (aa 1–826) is depicted on top; bHLH domain is represented by a black box, PAS domains labeled as PAS, and two PEST-like sequences drawn as hatched boxes. The Gal4 DNA binding domain (G4) is fused to stretches of HIF-1α downstream of the PEST-like sequences (aa 498, 530, 577, and 602–814) and to stretches of HIF-1α upstream of the PEST-like sequences (aa 98–603, 573, 518, 496, 400, and 363). The hypoxia inducibility (IND) of each construct is summarized on the right. (B) 293 cells were transfected with Gal-HIF-1α (98 to 573, 518, 496, and 400), labeled with [35S]methionine at 1% O2, and chased at 21% O2 for 0.5, 1, and 2 hr. Immunoprecipitation was carried out with anti-Gal4 antibody. Gal4-HIF-1α fusions are denoted by an arrow, and nonspecific bands by open arrowheads.
We first examined the region containing the PEST-like sequences and downstream thereof (aa 498–814; Fig. 2A). In contrast to HIF-1, these Gal4-HIF-1α fusions were constitutively expressed and were not induced by hypoxia, except for Gal4-HIF-1α(498–814) that showed slightly weaker expression under normoxia (summarized in Fig. 2A). Likewise, DNA binding of these constitutively expressed Gal4 fusions was not altered by changes of oxygen levels, nor by cycloheximide (data not shown), a protein synthesis inhibitor known to abrogate HIF-1 DNA binding (3).
Remarkably, HIF-1-like DNA binding was observed when the region of HIF-1α downstream from the PAS domain through the two PEST-like sequences (aa 98–603) was examined (Fig. 2A). In particular, Gal4-HIF-1α(98–603) containing both PEST-like sequences displayed a robust hypoxic induction (+++) as compared with HIF-1 binding (++++). The hypoxic induction became attenuated when one of the PEST-like sequences was deleted [Gal4- HIF-1α(98–573 and 98–518)]. Gal4-HIF-1α(98–496) still showed a weak induction even though both PEST-like sequences were removed, suggesting that the region controlling oxygen-dependent degradation was not limited to the PEST-like sequences. However, only constitutive binding (absence of induction) was obtained when Gal4-HIF-1α(98–400 and 98–363) were tested. Consistent with these results, cycloheximide inhibited DNA binding of Gal4-HIF-1α species that were induced by hypoxia, but not those constitutively expressed (data not shown). Pulse-chase experiments (Fig. 2B) confirmed that hypoxia-induced Gal4-HIF-1α(98–573, 98–518, and 98–496) had a faster turnover rate than that of Gal4-HIF-1α(98–400) under normoxia. Taken together, these data indicate that the oxygen regulatory region lies within aa 401 and 603 of HIF-1α and that acquisition of this region rendered Gal4, a constitutively expressed protein, unstable under normoxia.
To confirm and extend the above results, HIF-1α expression vectors with C-terminal deletions were constructed, transfected into 293 cells, and determined by Western blot analysis or immunoprecipitation. Because our anti-HIF-1α antibodies were made against the C terminus, an HA-tag was placed in frame at the N terminus so that the anti-HA antibody could recognize HIF-1α with C-terminal truncations. Consistent with the above results, Western blot analyses and immunoprecipitations revealed that C-terminal deletions to aa 653, 521, and 494 gave rise to gradually increasing, expression of HIF-1α under normoxia, whereas deletions to aa 400 and 329 resulted in strong constitutive expression without hypoxic induction (data not shown).
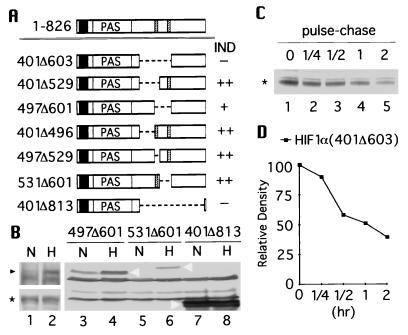
These results suggest that degradation of HIF-1α is governed by an oxygen-dependent degradation (ODD) domain that enables an oxygen-dependent signal to effect degradation of the protein. To directly test this hypothesis, the stretch of amino acids (401–603) was removed from HA-tagged HIF-1α (Fig. 3A). Western blot assays with the anti-HIF-1α antibodies showed that in contrast to the full-length HIF-1α (Fig. 3B Upper, lanes 1 and 2), HIF-1α(401Δ603) was constitutively expressed and indifferent to changes in oxygen tension (Lower, lanes 1 and 2). The use of antibodies that bind specifically to the C terminus of HIF-1α confirmed that the remaining C-terminal portion of HIF-1α(401Δ603) was expressed. Identical results were obtained with fusion construct HIF-1α(401Δ813) containing a larger internal deletion (Fig. 3B, lanes 7 and 8). These results suggest that aa 401–603 contains the ODD domain.
Figure 3.
Analyses of internal deletions of HIF-1α. (A) Schematic drawings of internal deletions of HIF-1α where aa 401–603, 401–529, 497–601, 401–496, 497–529, 531–601, and 401–813 were deleted. Hypoxic inducibility of each construct is summarized on the right. (B) HA-tagged full-length HIF-1α (Upper) and HIF-1α(401Δ603) (Lower) were cotransfected with pARNT into 293 cells and examined by Western blot analysis using anti-HIF-1α antibodies. Likewise, HA-tagged HIF-1α(497Δ601, 531Δ601, and 401Δ813) were cotransfected with pARNT and analyzed by Western blot using anti-HA antibody (lanes 3–8). (C) 293 cells were transfected with HA-tagged HIF-1α(401Δ603), labeled with [35S]methionine, and then chased for 0.25, 0.5, 1, and 2 hr under normoxia. Immunoprecipitation was done with anti-HA antibody. The intensity of each band was quantified and plotted in D. The full-length HIF-1α is marked with black arrow head, HIF-1α(401Δ603) with asterisk, and HIF-1α(497Δ601, 531Δ601, and 401Δ813) by white arrowhead.
An attempt was made to narrow down the ODD domain by dividing the aa 401–603 region into three parts (Fig. 3A): aa 401–496; aa 497–529 containing one PEST-like sequence; and aa 531–601 containing the other PEST-like sequence. Systematic internal deletions of each of these three parts and combinations thereof were tested to determine the minimum requirement for the ODD domain. Unlike HIF-1α(401Δ603), these partially deleted HIF-1α displayed various degrees of instability under normoxia (summarized in Fig. 3A; Fig. 3B Right). Thus, deletion of the entire ODD domain was necessary for complete stabilization of HIF-1α.
The stability of HIF-1α(401Δ603) was confirmed by pulse-chase (Fig. 3C) demonstrating that the half-life of the protein under normoxia was markedly increased to ≈1 hr (Fig. 3D). Together, these results confirm that the ODD domain lies between aa 401 and 603 of HIF-1α and is the critical determinant of oxygen-induced instability.
The ODD Domain Alone Confers ODD.
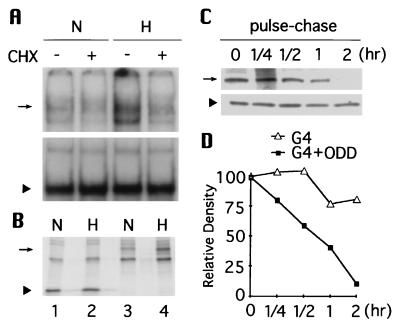
To determine if the ODD domain alone can function independently in a heterologous fashion, the aa 401–603 portion of HIF-1α was fused to Gal4. In contrast to the DNA binding pattern exhibited by Gal4 (Fig. 4A Lower), addition of the ODD domain resulted in a marked decrease in Gal4 DNA binding under normoxia (Fig. 4A Upper, lanes 1 and 3). Consistent with its blocking activation of HIF-1 (3), cycloheximide abrogated DNA binding of Gal4-HIF-1α(401–603), but not that of Gal4 (lanes 2 and 4). Protein abundance was also measured by immunoprecipitation (Fig. 4B). In keeping with the gel shift results, the expression of Gal4-HIF-1α(401–603) was considerably lowered under normoxia compared with that under hypoxia (lanes 3 and 4), whereas the expression of Gal4 was insensitive to oxygen (lanes 1 and 2). Furthermore, pulse-chase experiments (Fig. 4C) demonstrated that the ODD domain shortened the Gal4 half-life from far more than 2 hr to ≈45 min (Fig. 4D). Thus, the ODD domain is both necessary and sufficient for triggering O2-dependent degradation of HIF-1α. It is noteworthy that under normoxia the half-life of Gal4-HIF-1α(401–603) is much longer (45 min) than that of HIF-1α (<5 min). This difference is presumably attributable to the intrinsic stability of Gal4 protein (t1/2 ≫ 2 hr) vs. the relative instability of the ODD domain-deleted HIF-1α (t1/2 ≈ 1 hr).
Figure 4.
The function of ODD domain is transportable. (A) Gal4-HIF-1α(401–603) (Upper) and Gal4 (Lower) expression vectors were transfected into 293 cells respectively and their DNA-binding activity was tested by electrophoretic mobility-shift assay. (B) [35S]methionine-labeling and immunoprecipitation were performed to quantify abundance of Gal4 (lanes 1 and 2) and Gal4-HIF-1α(401–603) (lanes 3 and 4) under normoxia and hypoxia. (C) Similarly, Gal4-HIF-1α(401–603) (Upper) and Gal4 (Lower) were pulse-labeled and chased for 0.25, 0.5, 1, and 2 hr at 21% O2. The intensity of each signal was plotted in D. Arrow indicates Gal4-HIF-1α(401–603) and arrowhead marks Gal4.
Autonomous Heterodimerization, DNA-Binding, and Transactivation upon Removal of the Entire ODD Domain.
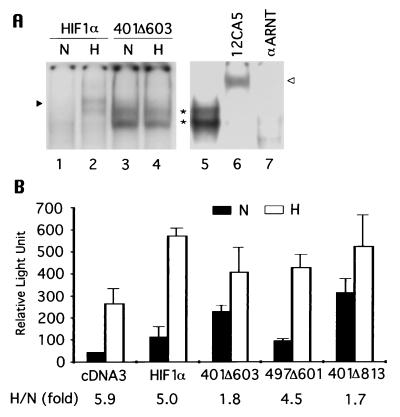
Because it is necessary for HIF-1α to heterodimerize with ARNT to bind to DNA, HIF-1α(401Δ603) was cotransfected with an ARNT expression vector into 293 cells to test its DNA binding ability. As shown in Fig. 5A, HIF-1α(401Δ603) was capable of binding to the hypoxic response element and did so equally well, if not better, at 21% O2 compared with 1% O2 (lanes 3 and 4). This binding was a result of spontaneous heterodimerization, as demonstrated by an electrophoretic mobility shift assay where HIF-1α(401Δ603)-DNA complexes were supershifted or abolished by the addition of antibodies against HA or ARNT (lanes 6 and 7). These results suggest that once HIF-1α is stabilized, heterodimerization and subsequent DNA binding do not require hypoxia signaling.
Figure 5.
Autonomous heterodimerization, DNA binding, and transactivation of stable HIF-1α. (A) (HA)HIF-1α or (HA)HIF-1α(401Δ603) were cotransfected with pARNT into 293 cells and HIF-1 DNA binding was examined directly by electrophoretic mobility-shift assay (lanes 1–4), or in the presence of antibodies against HA or ARNT (21) (lanes 6 and 7). An arrowhead denotes HIF-1, an asterisk indicates HIF-1α(401Δ603), and an open arrowhead marks supershifted HIF-1α(401Δ603). (B) Hep3B cells were transfected with pEpoE-luc and pcDNA3 or with pEpoE-luc and pARNT plus HA-tagged HIF-1α, HIF-1α(401Δ603), HIF-1α(497–601), or HIF-1α(401Δ813). Luciferase activity was measured and plotted. The results are the mean of three independent transfections. The corresponding hypoxic induction fold is indicated at bottom.
The transactivation domains of HIF-1α have been mapped to aa 540–580 and 780–826 (16, 17), and therefore HIF-1α(401Δ603) lacks the former but retains the latter. Given its autonomous DNA binding activity, HIF-1α(401Δ603) was tested for transactivation ability by cotransfecting Hep3B cells with an erythropoietin 3′ enhancer-driven reporter (21). As depicted in Fig. 5B, under normoxia reporter alone showed a background level of luciferase activity, which was markedly enhanced (≈6 fold) by hypoxia as a result of activation of the endogenous HIF-1. When the reporter was cotransfected with vectors overexpressing ARNT and the wild-type HIF-1α, luciferase activity was stimulated under both normoxia and hypoxia, resulting in a 5-fold hypoxic induction. In contrast, HIF-1α(401Δ603) particularly increased the reporter activity under normoxia, reducing hypoxic induction to <2-fold. Similar results were obtained with the larger deletion construct HIF-1α(401Δ813), which contains only 13 residues of the C-terminal transactivation domain. However, the hypoxic induction of the oxygen-sensitive HIF-1α(497Δ601) (Fig. 3B) was similar to that of full-length HIF-1α.
DISCUSSION
We postulated previously that activation of HIF-1 depends primarily on hypoxia-induced stabilization of HIF-1α, which is otherwise rapidly degraded in oxygenated cells (21). In this report, we identified an ODD domain through three independent approaches: Gal4-HIF-1α fusions for initial screening, HIF-1α C-terminal deletions for confirmation, and HIF-1α internal deletions for final verification. All the data are internally consistent, strongly supporting the functional importance of the ODD domain in controlling HIF-1α stability. In cells exposed to 21% O2, the ODD domain confers on full-length HIF-1α an extremely short half-life (≈5 min), even less than that of c-Fos, which is also regulated by the ubiquitin-proteasome pathway. Such rapid turnover is appropriate for tight regulation of transcription factors such as HIF-1α and c-Fos, enabling cells to adapt rapidly to changes in environmental stimuli.
There is growing evidence that ubiquitin-proteasome mediated degradation plays a critical role in the regulation of a wide range of transcription factors (for reviews see refs. 28–30). Our experiments demonstrate that this pathway mediates the oxygen-dependent proteolysis of HIF-1α, a transcription factor important in adaptation to hypoxia. Recently, Salceda and Caro (31) reported that heterodimeric HIF-1 DNA binding activity is enhanced by inhibition of ubiquitin-dependent proteolysis. However, no information was presented on which subunits of HIF-1 are involved in this process.
Proteins containing PEST sequences have a high probability of being targets for rapid intracellular degradation (26, 27). The human HIF-1α contains two PEST-like motifs at positions 499–518 and 581–600 (5). These sequences lie at the carboxyl end of the ODD domain, shown to be involved in O2-dependent degradation of HIF-1α. However, the N-terminal half of the ODD domain is also involved in degradation. This is suggested particularly by the data from HIF-1α C-terminal deletions and internal deletions. Most closely related to HIF-1α in the bHLH-PAS family is EPAS-1/HLF/MOP2 (32–34), which is selectively expressed in endothelial cells and may play a critical role in cell differentiation. While there are no sequences in EPAS-1/HLF/MOP2 that are homologous to the ODD domain, putative PEST motifs are present elsewhere in the protein, the function of which is unclear. In addition, the aryl hydrocarbon receptor bears a number of similarities to HIF-1α, but, unlike HIF-1α, lacks PEST motifs and is relatively stable, irrespective of its ligation state (35).
Phosphorylation may be critical in the activation of HIF-1 by hypoxia. Treatment of cells with inhibitors of both serine/threonine and tyrosine kinases block hypoxic induction of HIF-1 activation (18). It will be important to ascertain whether oxygen affects phosphorylation of HIF-1α at specific sites and to determine how this might impact on ubiquitin-proteasome-mediated degradation. Although the ODD domain is relatively large, partial internal deletions failed to give rise to a stable HIF-1α, indicating functional redundancy within this domain. Indeed, among the three artificially divided parts of the ODD domain (aa 401–496, 497–529, and 530–603), each of them independently conferred hypoxic induction to different degrees (Fig. 3A). These results pose a stiff challenge to determining the precise sites responsible for O2-dependent degradation. There are likely to be multiple functional sites spread over the domain so that HIF-1α would not be stabilized unless all or most of the functional sites have been mutated simultaneously. Recently Pugh et al. (17) examined the interior transactivation domain of HIF-1α (aa 549–582) that is within the ODD domain. Replacements of serine, threonine, and tyrosine residues in this stretch had no effect on hypoxic induction of a reporter.
Regulation of transcription factors by the ubiquitin-proteasome pathway may be dependent on interactions with other proteins. For example, when E2Fs are bound to pocket proteins (Rb, p107, and p130), they are protected from degradation (36, 37). Conversely, proteasome-mediated degradation of c-Fos is enhanced by the presence of c-Jun (38). These observations raise questions of how heterodimerization with ARNT may impact on the stability of HIF-1α. Binding to full-length ARNT protects HIF-1α from in vitro proteolytic degradation (20). Although we did notice that cotransfection with ARNT and the ODD domain-deleted HIF-1α(401Δ603) resulted in enhancement of DNA binding activity as compared with those transfected with HIF-1α(401Δ603) alone (data not shown), ARNT did not further increase DNA binding (Fig. 5A) or protein abundance (Fig. 3B) of HIF-1α(401Δ603) under hypoxia. ARNT may play a role in vivo for stabilizing HIF-1α upon dimerization, but in an oxygen-independent manner. Moreover, this protective role of ARNT may be limited by the fact that HIF-1α decays rapidly upon switching cells from 1% O2 to 21%, despite its dimerization with constitutively expressed endogenous ARNT (21).
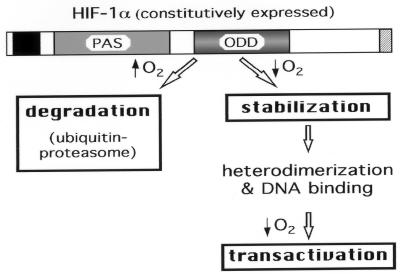
The identification of the ODD domain has provided a molecular basis for understanding the mechanism by which HIF-1 is activated. As depicted in Fig. 6, it is likely that the activation process of HIF-1 by hypoxia involves the following three sequential steps. (i) Hypoxia stabilizes HIF-1α by abrogating the ODD domain-mediated ubiquitin-proteasome degradation; (ii) stabilized HIF-1α autonomously heterodimerizes with endogenous ARNT and binds to the hypoxic response element; and (iii) hypoxia enhances HIF-1 transactivation of target genes. Stabilization of HIF-1α is arguably the primary mode of regulation because the other two steps depend on the abundance of HIF-1α protein. Moreover, the ODD domain may serve as a starting point for delineating the hypoxic signaling pathway, enabling identification of proteins that interact with this region. Furthermore, given the transportability of ODD domain function, it is conceivable that ODD fusion proteins can be engineered so that the expressed protein of interest could be regulated in a physiological system by oxygen tension.
Figure 6.
A model for hypoxia-induced activation of HIF-1. A schematic drawing of HIF-1α is illustrated. The bHLH domain is marked by a black box and the C terminus transactivation domain by a hatched box. PAS and ODD domains are indicated. Steps regulated by changes of oxygen tension are boxed with bold lines.
Acknowledgments
We thank Zoltan Arany, Shoumo Bhattacharya and David M. Livingston for anti-HIF-1α antibodies and the Gal4 expression vector, Dirk Bohmann for the HA-tagged ubiquitin construct, and Oliver Hankinson for the ARNT cDNA clone. We also thank Alfred Goldberg for helpful discussions. This work was supported by National Institutes of Health Grant RO1-DK41234 (H.F.B.). L.E.H. is a recipient of the National Research Service Award from the National Institutes of Health (F32-DK09365).
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: HIF-1, hypoxia-inducible factor 1; ODD, oxygen-dependent degradation; Cbz-LLL, N-carbobenzoxyl-l-leucinyl-l-leucinyl-l-norvalinal; ARNT, aryl hydrocarbon nuclear translocator; bHLH, basic helix-loop-helix; HA, hemagglutinin; PEST, proline, glutamic acid, serine, threonine; β-gal, β-galactosidase..
References
- 1.Bunn H F, Poyton R O. Physiol Rev. 1996;76:839–885. doi: 10.1152/physrev.1996.76.3.839. [DOI] [PubMed] [Google Scholar]
- 2.Guillemin K, Krasnow M A. Cell. 1997;89:9–12. doi: 10.1016/s0092-8674(00)80176-2. [DOI] [PubMed] [Google Scholar]
- 3.Semenza G L, Wang G L. Mol Cell Biol. 1992;12:5447–5454. doi: 10.1128/mcb.12.12.5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang G L, Semenza G L. J Biol Chem. 1993;268:21513–21518. [PubMed] [Google Scholar]
- 5.Wang G L, Jiang B-H, Rue E A, Semenza G L. Proc Natl Acad Sci USA. 1995;92:5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoffman E C, Reyes H, Chu F-F, Sander F, Conley L H, Brooks B A, Hankinson O. Science. 1991;252:954–958. doi: 10.1126/science.1852076. [DOI] [PubMed] [Google Scholar]
- 7.Reisz-Porszasz S, Probst M, Fukunaga B, Hankinson O. Mol Cell Biol. 1994;14:6075–6086. doi: 10.1128/mcb.14.9.6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang B-H, Rue E, Wang G L, Roe R, Semenza G L. J Biol Chem. 1996;271:17771–17778. doi: 10.1074/jbc.271.30.17771. [DOI] [PubMed] [Google Scholar]
- 9.Huang Z J, Edery I, Rosbach M. Nature (London) 1993;364:259–262. doi: 10.1038/364259a0. [DOI] [PubMed] [Google Scholar]
- 10.Lindebro M C, Poellinger L, Whitelaw M L. EMBO J. 1995;14:3528–3539. doi: 10.1002/j.1460-2075.1995.tb07359.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zelzer E, Wappner P, Shilo B-Z. Genes Dev. 1997;11:2079–2089. doi: 10.1101/gad.11.16.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jain S, Dolwick K M, Schmidt J V, Bradfield C A. J Biol Chem. 1994;269:31518–31524. [PubMed] [Google Scholar]
- 13.Li H, Dong L, Whitlock J P. J Biol Chem. 1994;269:28098–28105. [PubMed] [Google Scholar]
- 14.Wood S M, Gleadle J M, Pugh C W, Hankinson O, Ratcliffe P J. J Biol Chem. 1996;269:15117–15123. doi: 10.1074/jbc.271.25.15117. [DOI] [PubMed] [Google Scholar]
- 15.Li H, Ko H P, Whitlock J P. J Biol Chem. 1996;271:21262–21267. doi: 10.1074/jbc.271.35.21262. [DOI] [PubMed] [Google Scholar]
- 16.Jiang B-H, Zheng J Z, Leung S W, Roe R, Semenza G L. J Biol Chem. 1997;272:19253–19260. doi: 10.1074/jbc.272.31.19253. [DOI] [PubMed] [Google Scholar]
- 17.Pugh C W, O’Rourke J F, Nagao M, Gleadle J M, Ratcliffe P J. J Biol Chem. 1997;272:11205–11214. doi: 10.1074/jbc.272.17.11205. [DOI] [PubMed] [Google Scholar]
- 18.Wang G L, Jiang B-H, Semenza G L. Biochem Biophys Res Commun. 1995;216:669–675. doi: 10.1006/bbrc.1995.2674. [DOI] [PubMed] [Google Scholar]
- 19.Gradin K, McGuire J, Wenger R H, Kvietikova I, Whitelaw M, Toftgard R, Tora L, Gassman M, Poellinger L. Mol Cell Biol. 1996;16:5221–5231. doi: 10.1128/mcb.16.10.5221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kallio P J, Pongratz I, Gradin K, McGuire J, Poellinger L. Proc Natl Acad Sci USA. 1997;94:5667–5672. doi: 10.1073/pnas.94.11.5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang L E, Arany Z, Livingston D M, Bunn H F. J Biol Chem. 1996;271:32253–32259. doi: 10.1074/jbc.271.50.32253. [DOI] [PubMed] [Google Scholar]
- 22.Bhattacharya S, Eckner R, Grossman S, Oldread E, Arany Z, D’Andrea A, Livingston D M. Nature (London) 1996;383:344–347. doi: 10.1038/383344a0. [DOI] [PubMed] [Google Scholar]
- 23.Gu J, Parthasarathi S, Varela-Echavarria A, Ron Y, Dougherty J P. Virology. 1995;206:885–893. doi: 10.1006/viro.1995.1011. [DOI] [PubMed] [Google Scholar]
- 24.Rock K, Gramm C, Rothstein L, Clark K, Stein R, Dick L, Hwang D, Goldberg A. Cell. 1994;78:761–771. doi: 10.1016/s0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- 25.Wang G L, Semenza G L. Blood. 1993;82:3610–3615. [PubMed] [Google Scholar]
- 26.Rogers S, Wells R, Rechsteiner M. Science. 1986;234:364–368. doi: 10.1126/science.2876518. [DOI] [PubMed] [Google Scholar]
- 27.Rechsteiner, M. & Rogers, S. W. (1996) Trends Biochem. Sci. 21. [PubMed]
- 28.Ciechanover A. Cell. 1994;79:13–21. doi: 10.1016/0092-8674(94)90396-4. [DOI] [PubMed] [Google Scholar]
- 29.Coux O, Tanaka K, Goldberg A L. Annu Rev Biochem. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- 30.Hilt W, Wolf D H. Trends Biochem Sci. 1996;21:96–102. [PubMed] [Google Scholar]
- 31.Salceda S, Caro J. J Biol Chem. 1997;272:22642–22647. doi: 10.1074/jbc.272.36.22642. [DOI] [PubMed] [Google Scholar]
- 32.Tian H, McKnight S L, Russell D W. Genes Dev. 1997;11:72–82. doi: 10.1101/gad.11.1.72. [DOI] [PubMed] [Google Scholar]
- 33.Ema M, Taya S, Yokotani N, Sogawa K, Matsuda Y, Fujii-Kuriyama Y. Proc Natl Acad Sci USA. 1997;94:4273–4278. doi: 10.1073/pnas.94.9.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hogenesch J B, Chan W K, Jackiw V H, Brown R C, Gu Y-Z, Pray-Grant M, Perdew G H, Bradfield C A. J Biol Chem. 1997;272:8581–8593. doi: 10.1074/jbc.272.13.8581. [DOI] [PubMed] [Google Scholar]
- 35.Hankinson O. Annu Rev Pharmacol Toxicol. 1995;35:307–340. doi: 10.1146/annurev.pa.35.040195.001515. [DOI] [PubMed] [Google Scholar]
- 36.Hateboer G, Kerkhoven R M, Shvarts A, Bernards R, Beijersbergen R L. Genes & Dev. 1996;10:2960–2970. doi: 10.1101/gad.10.23.2960. [DOI] [PubMed] [Google Scholar]
- 37.Hoffman F, Martelli F, Livingston D M, Wang Z. Genes & Dev. 1996;10:2949–2959. doi: 10.1101/gad.10.23.2949. [DOI] [PubMed] [Google Scholar]
- 38.Tsurumi C, Ishida N, Tamura T, Kakizuka A, NIshida E, Okumura E, Kishimoto T, Inagaki M, Okazaki K, Sagata N, Ichihara A, Tanaka K. Mol Cell Biol. 1995;15:5682–5687. doi: 10.1128/mcb.15.10.5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee D H, Goldberg A L. J Biol Chem. 1996;271:27280–27284. doi: 10.1074/jbc.271.44.27280. [DOI] [PubMed] [Google Scholar]