Abstract
Arsenite, the trivalent form of arsenic present in the environment, is a known human carcinogen that lacked mutagenic activity in bacterial and standard mammalian cell mutation assays. We show herein that when evaluated in an assay (AL cell assay), in which both intragenic and multilocus mutations are detectable, that arsenite is in fact a strong dose-dependent mutagen and that it induces mostly large deletion mutations. Cotreatment of cells with the oxygen radical scavenger dimethyl sulfoxide significantly reduces the mutagenicity of arsenite. Thus, the carcinogenicity of arsenite can be explained at least in part by it being a mutagen that depends on reactive oxygen species for its activity.
Environmental carcinogens have been postulated to be an important etiological factor for the majority of human cancer (1). A better understanding of the carcinogenic mechanisms of these agents will facilitate not only the development of treatment modalities but prevention strategies as well. Arsenic, a known human carcinogen and teratogen, is ubiquitously present in the environment, where it occurs mainly as compounds of arsenite (As3+) and arsenate (As5+). Epidemiological studies have shown that chronic exposure to arsenic can result in liver injury, peripheral neuropathy, and an increased incidence of cancer of the lung, skin, bladder, and liver (1). In addition, occupational exposure to arsenic increases the lung cancer incidence among smokers and underground miners in a manner consistent with a synergistic interaction of arsenic with tobacco smoke or radon found in mines (2, 3). Animal models are of little help in understanding its mechanism; for reasons that are not clear arsenic is not carcinogenic in rodent models. It is one of the few, possibly the only, well-established human carcinogen that does not induce tumors in laboratory animals (4, 5).
In the absence of animal models, in vitro studies become particularly important in providing information on the carcinogenic mechanisms of arsenic. Arsenic and arsenical compounds are toxic to and induce morphological transformants in Syrian hamster embryo and murine C3H 10T½ cells (6, 7). These agents are also potent clastogens both in vivo and in vitro. They can, for example, induce sister chromatid exchanges and chromosome aberrations in both human and rodent cells in culture (4, 6, 8, 9) and in cells of exposed humans (10). Arsenical compounds have also been shown to induce gene amplification, arrest cells in mitosis, inhibit DNA repair, and induce expression of the c-fos gene and the oxidative stress protein heme oxygenase in mammalian cells (11–13), and they have been implicated as promoters and comutagens for a variety of agents (14). In light of these actions, it has been puzzling that arsenic is only weakly active or, more often, completely inactive in bacterial and mammalian cell mutation assays (4–6, 15–17). One plausible explanation is that arsenic induces mostly multilocus deletions that are incompatible with cell survival when mutations are measured at gene loci that are closely linked to essential genes. In other words, it is possible that many of the types of mutations induced by arsenic are poorly recovered in these assays because they are lethal. An elucidation of the genotoxic effects of arsenic promises to shed light on mechanisms underlying its carcinogenic effects in humans, as well as its interaction with other environmental carcinogens such as tobacco smoke and radon. Furthermore, these studies may provide a model for the genotoxic actions of other heavy metals as well.
Toward this end we have quantified the induction of S1− and HPRT− mutants in human–hamster AL cells treated with graded doses of sodium arsenite and compared the types of mutation induced with those arising spontaneously. The human–hamster hybrid (AL) cells contain a standard set of hamster chromosomes plus a single human chromosome 11, which encodes a series of human cell surface antigens (18). One such antigen, S1¶ encoded by the MIC1 gene at 11p13.5, is especially useful in mutation studies, because its presence or loss as a result of mutation can easily be determined in a complement-mediated cytotoxicity assay (19–21). Because only a small part of region 11p15.5 is required for the viability of AL cells, mutations in the human chromosome 11 ranging in size up to 140 million bases of DNA can be detected. We report herein that when evaluated at the S1 locus arsenite is a significant mutagen that induces mainly large chromosomal mutations and that the mutagenic response is mediated by reactive oxygen species (ROS).
MATERIALS AND METHODS
Cell Culture.
The AL hybrid cells that contain a standard set of CHO-K1 chromosomes and a single copy of human chromosome 11 were used. Chromosome 11 encodes cellular surface markers that render AL cells sensitive to killing by specific monoclonal antibodies in the presence of rabbit serum complement (HPR, Denver, PA). Antibody E7.1 specific to the S1 (CD59) antigen was produced from hybridoma culture as described (19, 22). Cells were maintained in Ham’s F-12 medium supplemented with 8% heat-inactivated fetal bovine serum, gentamycin (25 μg/ml), and 2× normal glycine (2 × 10−4 M) at 37°C in a humidified 5% CO2/95% air incubator and passaged as described (20, 21, 23).
Determination of Arsenic Cytotoxicity.
A stock solution of sodium arsenite (Sigma) at 1 mg/ml was prepared in doubled-distilled water and sterilized by passing through a 0.22-μm (pore size) syringe filter. Working concentrations were prepared by diluting the stock with complete F-12 medium. To determine the dose–response of AL cells to arsenite, exponentially growing cells were trypsinized, replated into 25-cm2 tissue culture flasks at a density of 1 × 105 cells per flask, and treated 48 hr after plating with arsenite for either 1 or 5 days. After treatment, cultures were washed twice with balanced salt solution, trypsinized, and replated into 100-mm diameter Petri dishes for colony formation. Cultures were incubated for 7–12 days, at which time they were fixed with formaldehyde and stained with Giemsa. The number of colonies was counted to determine the surviving fraction as described (20, 23).
Mutation Assay.
S1 locus. After treatment, cultures were replated into T25 flasks and cultured for 7 days. This expression period is needed to permit surviving cells to recover from the temporary growth lag caused by arsenite and to multiply sufficiently so that the progeny of the mutated cells are no longer expressing lethal amounts of the S1 surface antigen. To determine mutant fractions, 5 × 104 cells per dish were plated into six 60-mm dishes in a total of 2 ml of growth medium as described (20, 21, 23). The cultures were incubated for 2 hr to allow for cell attachment, after which 0.3% S1 antiserum and 1.5% (vol/vol) freshly thawed complement were added to each dish. After overnight incubation, this medium was removed, and the cultures were further incubated in standard growth medium for 8 days. At this time the cells were fixed and stained, and the number of S1− mutant colonies was scored. Controls included identical sets of dishes containing antiserum alone, complement alone, or neither agent. The cultures derived from each treatment dose were tested for mutant yield for two consecutive weeks to ensure full expression of the mutations. The mutant fraction at each dose (MF) was calculated as the number of surviving colonies divided by the total number of cells plated after correction for any nonspecific killing due to complement alone. The mutant yield (MY) is the slope of the dose–response curve and is independent of the background mutant level.
HPRT locus. Aliquots of treated AL cells were plated at 2 × 105 cells per dish in 10 ml of medium containing 40 μM 6-thioguanine as described (20, 23). Corresponding dishes were plated at lower cell density in normal medium to determine plating efficiencies. Mutant frequencies were expressed as the number of mutants per 105 survivors.
Treatment with Dimethyl Sulfoxide (DMSO).
To examine the role of ROS in arsenite mutagenesis, exponentially growing AL cells were exposed to arsenite for 24 hr with or without concurrent treatment with graded doses of DMSO ranging from 0.05 to 0.25%. DMSO at the doses used was nontoxic and nonmutagenic and has been shown in a variety of in vitro and in vivo studies to be an effective free radical scavenger (24, 25). After treatment, cultures were washed, trypsinized, and replated for both survival and mutagenesis as described above.
Analysis of Mutant Spectrum by Multiplex PCR.
Independently derived S1− mutants were isolated by cloning and expanded in culture as described (21, 23). To ensure their clonal origin, either a single colony or, at times, two well-separated colonies per culture dish were isolated. A minimum of 25 mutants from each treatment group and more than 50 spontaneous mutants were analyzed. DNA was extracted by a salting-out procedure (21, 23).
Five DNA markers on chromosome 11 (Wilms’ tumor, parathyroid hormone, catalase, RAS, and apolipoprotein A-1) were chosen for multiplex PCR analysis because of their map positions relative to the MIC1 gene, which encodes the S1 antigen (18, 19, 22), and the availability of PCR primers for the coding regions of these genes (26–28). PCR amplifications were performed for 30 cycles using a DNA thermal cycler model 480 (Perkin–Elmer/Cetus) in 20-μl reaction mixtures containing 0.2 μg of the EcoRI-digested DNA sample in 1× Stoffel fragment buffer, all four dNTPs (each at 0.2 mM), 3 mM MgCl2, 0.2 mM each primer, and 2 units of Stoffel fragment enzyme (21, 29). Each PCR cycle consisted of denaturation at 94°C for 1 min, annealing at 55°C for 1 min, and extension at 72°C for 1 min. After the last cycle, the samples were incubated at 72°C for an additional 20 min, electrophoresed on 2% agarose gels, and stained with ethidium bromide.
RESULTS
Toxicity of Arsenite to AL Cells.
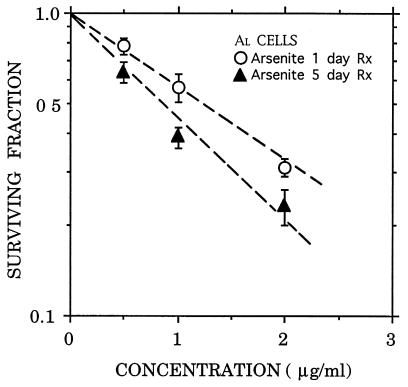
Arsenite induced a dose-dependent toxicity in AL cells as shown in Fig. 1, where the surviving fractions, after either a 1- or a 5-day continuous exposure, are plotted against drug concentration. The survival data fit well to a log–linear curve, and the lack of a shoulder in the dose–response curves is consistent with previous reports that arsenic inhibits DNA repair or recovery processes that normally act to produce a shoulder (15, 30). The mean lethal doses, Do, defined as the concentrations that reduced survival to 0.37 (1/e) in the log–linear portion of the curves for the 5- and 1-day treatments, were 1.3 and 1.7 μg/ml, respectively. These curves were used to select appropriate doses for mutation analysis.
Figure 1.
Survival curve for AL cells exposed to graded doses of sodium arsenite. Exponentially growing AL cells were treated with the drug for either 1 or 5 days. Each data point represents an average of four experiments. The Do values of the curves were 1.7 and 1.3 μg/ml, respectively. Error bars represent ±SEM.
Mutagenicity of Arsenite.
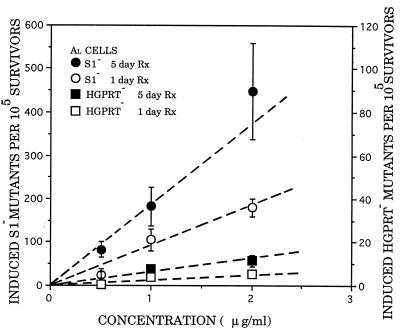
Dose–response curves for the induction of HPRT− and S1− mutants by arsenite are shown in Fig. 2, where the induced mutant yields (background subtracted) per 105 survivors are plotted against arsenite concentration. The curves were fitted by using the method of least squares. Consistent with previous studies (4–6), arsenite induced relatively few HPRT− mutants over the range of doses examined. The 1-day treatment gave a slight but dose-dependent increase in HPRT− mutants with a slope (MY) of 2.5 mutants per 105 clonogenic survivors per μg per ml, 35-fold less than the MY = 88 for induction of S1− mutants. The frequency of spontaneous HPRT− mutants ranged from 0.8 to 5.4 per 105 clonogenic survivors, whereas the S1− locus averaged 50 per 105 survivors. Mutant fraction was determined at 7–14 days after exposure to arsenite, and the incidence remained fairly constant over the assay period. A perspective on this mutagenic potency is provided by comparing the mutagenicity of arsenite with that of known clastogenic carcinogens such as γ-rays and chrysotile asbestos. Expressed in terms of mutant fractions at the DO dose, which we have previously shown on practical and theoretical grounds to be a particularly relevant comparison (19, 20, 29), arsenite and ionizing radiation were about equally mutagenic: the MF per DO for arsenite was 140 compared with 130 for γ-rays (19) and 165 for asbestos fibers (23).
Figure 2.
Mutation induction in AL cells by graded doses of sodium arsenite, expressed as number of induced mutants per 105 clonogenic survivors, at the S1 and HPRT loci after either 1- or 5-day treatment. Induced mutant fractions are the total mutant yield minus background. Each point represents data pooled from three to five experiments. Error bars represent ±SEM.
Mutagenicity of Arsenite Depends on Treatment Time.
We found that mutant induction by arsenite depended on the length of the treatment period. For example, the MY for S1− mutants for a 5-day treatment was 184 (Fig. 2) compared with 88 for a 1-day exposure. Similarly, the MY for HPRT− mutants increased from 2.5 at 1 day to 6.5 after a 5-day treatment period. This represents a 28-fold difference in mutant yield between the S1 and HPRT loci for a 5-day treatment.
Analysis of Mutant Spectra.
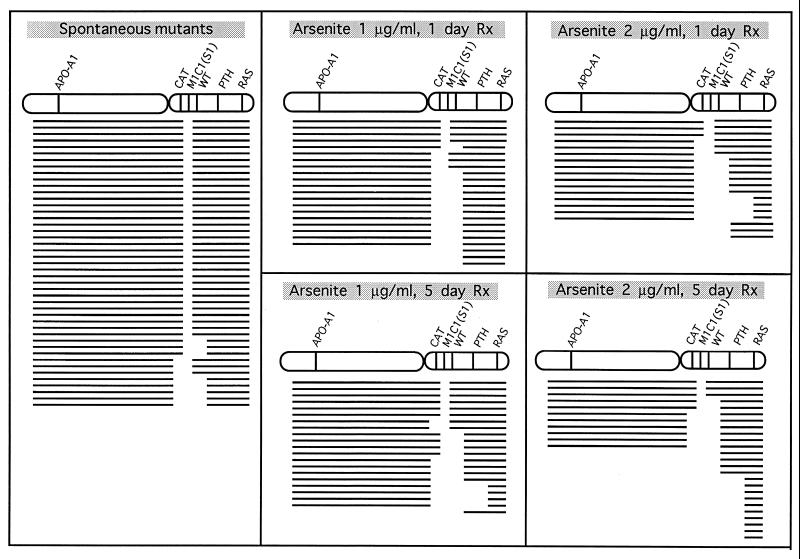
To determine the types of mutation that cause the S1− phenotype in arsenite-treated AL cells, we isolated individual clones and applied multiplex PCR to determine the presence or absence of five chromosome 11 markers located on either side of the MIC1 gene (Fig. 3). These primers and PCR conditions were selected to amplify only the human genes and not their CHO cognates (21, 29, 30). Because AL cells have only one chromosome 11, the presence or absence of the corresponding PCR products indicates that a particular segment of DNA containing these genes is present or missing, respectively. Previous studies have shown that a small region of the distal end of human chromosome 11 at 11p15.5 is required for survival of the AL cells (30). The obligate presence of this region identified herein by the RAS probe in all mutants provides a convenient internal PCR control. As shown in Fig. 4, the majority of spontaneous S1− mutants (31 of 42 mutants or 74%) retained all five of the chromosome 11 markers, whereas only 16 of 88 mutants (18%) from arsenic-treated cells were of this type. These values are significantly different (P < 0.01). Thus, most of the S1− mutants (82%) from arsenic-treated cells have suffered deletion mutations of >3.6 million base pairs (distance between the CAT and the WT genes, Fig. 3) compared with 26% of spontaneous mutants. Furthermore, the proportion of mutants suffering loss of additional chromosomal markers increased with treatment period so that at 5 days 10 of 25 mutants or 40% of the mutants from the cells treated with 2 μg/ml had lost all four markers examined that spanned both arms of the human chromosome 11.
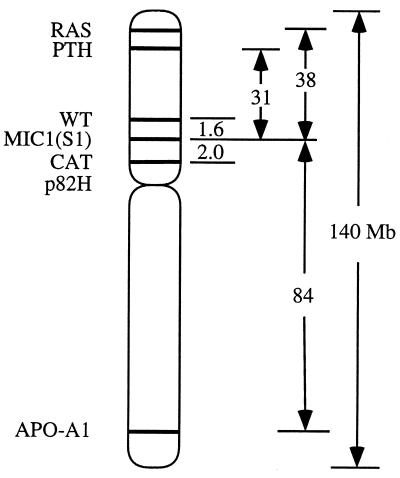
Figure 3.
Diagram of human chromosome 11 showing the M1C1 gene used in defining the S1− phenotype and the relative positions of other markers used in the multiplex PCR analysis to determine the extent of the S1 mutations. The M1C1 gene maps to 11p13.5. The two nearest markers flanking M1C1, CAT and WT, are separated by approximately 3.6 megabase pairs (Mbp) so that the S1− mutants that retained these neighboring markers could result from a base change to deletions as large as 3.6 Mbp.
Figure 4.
Deletion spectra of S1− mutants of spontaneous origin or from cells exposed to graded doses of sodium arsenite for either a 1- or a 5-day treatment period. Each line depicts the spectrum of a single independent mutant. The absence or presence of markers among the mutants was determined by multiplex PCR using DNA from S1− mutants as templates and primers for parathyroid hormone (PTH), Wilms’ tumor (WT), catalase (CAT), apolipoprotein A1 (APO-A1), and RAS. Blank space shows missing markers.
Mutagenesis of Arsenite Is Mediated by Oxyradicals.
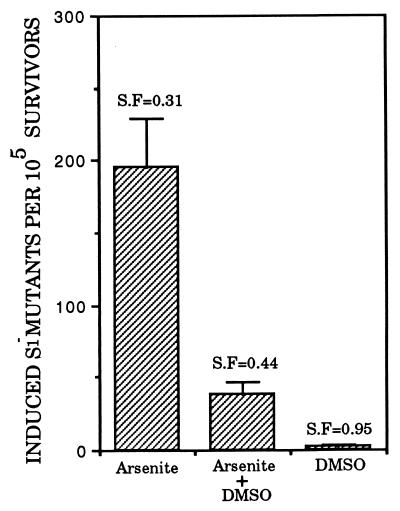
ROS such as superoxide anion, hydroxyl radicals, and hydrogen peroxides are the intermediates formed during oxidative metabolism. There is recent evidence to suggest that ROS mediate the dose-dependent increase in micronuclei in arsenite-treated CHO cells in vitro (31). Fig. 5 shows the suppressive effects of 0.1% DMSO on S1 mutant fraction induced by a concurrent 1-day treatment with arsenite at 2 μg/ml. DMSO alone was neither toxic nor mutagenic at the doses used (Fig. 5). Arsenite alone gave an induced S1− mutant yield of 195.5 ± 33 × 10−5 compared with 38.5 ± 7.7 × 10−5 when 0.1% DMSO was present (P < 0.01). A significant reduction in mutant yield was also observed with 0.05 or 0.25% DMSO (data not shown).
Figure 5.
Effects of the free radical scavenger DMSO (0.1%) on induced mutant yield in AL cells treated with sodium arsenite (2 μg/ml) for 24 hr. The survival fraction of the various treatment group is shown above each bar. Data were pooled from two or three experiments. Error bars represent ±SEM.
DISCUSSION
As a naturally occurring metalloid, arsenic is ubiquitously present in the environment. Epidemiological data gathered for more than a century have shown that arsenic is a potent human carcinogen. However, mechanisms by which arsenic induces cancer are not understood. The lack of suitable animal models necessitates reliance on in vitro studies to determine the cellular and molecular pathways involved. Previous genotoxic studies of arsenic have largely yielded negative findings for gene mutations but positive results for chromosomal aberrations (4, 6, 15, 16). The failure of arsenic to induce gene mutations in mammalian cells has been taken as evidence that a nongenotoxic pathway is responsible for arsenic-induced cancer through either hypomethylation of DNA (32) or inhibition of DNA ligation (33). Herein we show that arsenic is indeed mutagenic to endogenous genes in mammalian cells and that it induces mostly large multilocus deletions that are mediated through ROS.
Although human chromosome 11 in the AL hybrid resides in a hamster cell, there is no evidence that its function or structure is different than in its human cell habitat. Banding patterns for the human chromosome 11 in the AL cells are indistinguishable from any human chromosome number 11 (34). Furthermore, there is no evidence that the high frequencies of S1− mutations induced by a variety of mutagens such as radiation that induce predominately multilocus deletions are caused by the intrinsic hypermutability of the AL cell (19, 20, 29). For example, its mutability at the HPRT locus is no different from that of other human or rodent cell lines (19, 35). The main reason that more S1− mutants than ouabain-resistant or HPRT− mutants are obtained is that mutants with large deletions at the HPRT locus are poorly recovered. On the other hand, a deletion that inactivates the ouabain binding site that is required for generating ouabain-resistant mutants also deletes the ATP binding sites thereby rendering the mutants ouabain sensitive because the enzyme ATPase also cannot bind ATP and is, therefore, inactive (36).
In this regard, we recovered no ouabain-resistant mutants (3 mM ouabain) from arsenite-treated AL cell population (data not shown). The finding of induction of large mutations is consistent with the ample body of data on induction by arsenic of chromosome aberrations and micronuclei and with the recent report that arsenic induced both small and large colony mutants in the mouse lymphoma TK± (37). Thus, it appears that when studied in mutation assays that detect large chromosomal mutations, arsenic is in fact a potent mutagen/clastogen. Our result implicates ROS, especially hydroxyl radicals, as mediators of arsenite mutagenicity. These data are consistent with the recent demonstration that antioxidant enzymes such as superoxide dismutase, glutathione peroxidase, and particularly catalase reduce the incidence of arsenite-induced chromosomal aberrations and sister chromatid exchanges in cultured mammalian cells (31, 38). Furthermore, generation of intracellular oxidants in arsenite-treated porcine endothelial cells was prevented by treatment with the antioxidant N-acetylcysteine (39). Our present findings provide direct collaborating evidence on the role of ROS in mediating the mutagenic activities of arsenite.
The question arises that if the mutagenicity of arsenic is, in fact, mediated by ROS, why have gene mutation assays been negative after arsenic treatment? Part of the explanation is that although ROS can induce some intragenic mutations in bacteria and in mammalian cells (40, 41), they also induce large multilocus deletions and chromosome aberrations (42, 43) that would largely escape detection in HPRT and OUA. Also insufficient treatment time could be a factor in previously failed attempts to detect induced mutants in mammalian cells (4–6, 15, 16) because we show that the effect is time dependent. In our preliminary studies, we found that pretreatment of AL cells with buthionine (S,R)-sulfoximine, which depletes intracellular glutathione to less than 5% of control value, significantly enhanced the mutagenic potential of arsenite. Our results clearly demonstrate, however, that arsenite is a mutagen in cultured mammalian cells with normal levels of glutathione and that its mutagenicity, like its lethal and clastogenic effects, depends on ROS. Thus mutagenicity could underlie its carcinogenic activity.
Acknowledgments
We thank Drs. Eric Hall, Gloria Calaf, and Dan Gustafson, Ms. Diane Vannais, and Megan McGraw for valuable comments. This work was supported by National Institutes of Health Grants ES 08821, CA 49062, CA 36447, and CA 56392 and National Aeronautics and Space Administration Grant NAGW 4924.
ABBREVIATIONS
- DMSO
dimethyl sulfoxide
- ROS
reactive oxygen species
Footnotes
S1 antigen is now known to be the same as the CD59 antigen.
References
- 1.IARC Monograph on the Evaluation of the Carcinogenic Risk of Chemicals to Humans (1982) Suppl. 4, 50–51.
- 2.Hertz-Picciotto I, Smith S H, Holtzman D, Lipsett M, Alexeeff G. Epidemiology. 1992;3:23–31. doi: 10.1097/00001648-199201000-00006. [DOI] [PubMed] [Google Scholar]
- 3.Kusiak R A, Ritchie A C, Muller A C, Springer J. Br J Ind Med. 1993;50:920–927. doi: 10.1136/oem.50.10.920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barrett J C, Lamb P W, Wang T C, Lee T C. Biol Trace Elem Res. 1989;21:421–429. doi: 10.1007/BF02917284. [DOI] [PubMed] [Google Scholar]
- 5.Wang Z, Rossman T G. In: Toxicology of Metals. Cheng P, editor. Boca Raton, FL: CRC; 1996. pp. 221–229. [Google Scholar]
- 6.Lee T C, Oshimura M, Barrett J C. Carcinogenesis. 1985;6:1421–1426. doi: 10.1093/carcin/6.10.1421. [DOI] [PubMed] [Google Scholar]
- 7.Landolph J. Biol Trace Elem Res. 1989;21:459–467. doi: 10.1007/BF02917289. [DOI] [PubMed] [Google Scholar]
- 8.Nakamuro K, Sayato Y C. Mutat Res. 1981;88:73–80. doi: 10.1016/0165-1218(81)90091-4. [DOI] [PubMed] [Google Scholar]
- 9.Natarajan, A. T., Boei, J. J., Darroudi, F., Van Diemen, P. C., Doulout, F., Hande, M. P. & Ramalho, A. T. (1996) Environ. Health Perspect. 104 (Suppl. 3), 445–458. [DOI] [PMC free article] [PubMed]
- 10.Gonsebatt M E, Vega L, Salazar A M, Montero R, Guzman P, Blas J, Del Razo L M, Garcai-Vargas G, Albores A, Cebrian M E, Kelsh M, Ostrosky-Wegman P. Mutat Res. 1997;386:219–228. doi: 10.1016/s1383-5742(97)00009-4. [DOI] [PubMed] [Google Scholar]
- 11.Lee T C, Tanaka N, Lamb P W, Gilmer T M, Barrett J C. Science. 1988;241:79–81. doi: 10.1126/science.3388020. [DOI] [PubMed] [Google Scholar]
- 12.Vega L, Gonsebatt M E, Ostrosky-Wegman P. Mutat Res. 1995;334:365–373. doi: 10.1016/0165-1161(95)90074-8. [DOI] [PubMed] [Google Scholar]
- 13.Keyse S M, Applegate L A, Tromvoukis Y, Tyrell R M. Mol Cell Biol. 1990;10:4967–4972. doi: 10.1128/mcb.10.9.4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cavigelli M, Li W W, Lin A, Su B, Yushioka K, Karin M. EMBO, J. 1996;15:6269–6279. [PMC free article] [PubMed] [Google Scholar]
- 15.Rossman T G, Stone D, Molina M, Troll W. Environ Mutagen. 1980;2:371–379. doi: 10.1002/em.2860020307. [DOI] [PubMed] [Google Scholar]
- 16.Oberly T J, Piper C E, McDonald D S. J Toxicol Environ Mutagen. 1982;9:367–376. doi: 10.1080/15287398209530170. [DOI] [PubMed] [Google Scholar]
- 17.Yang J L, Chen M F, Wu C W, Lee T C. Environ Mol Mutagen. 1992;20:156–164. doi: 10.1002/em.2850200304. [DOI] [PubMed] [Google Scholar]
- 18.Puck T T, Wuchier P, Jones C, Kao F T. Proc Natl Acad Sci USA. 1971;68:3102–3106. doi: 10.1073/pnas.68.12.3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Waldren C A, Jones C, Puck T T. Proc Natl Acad Sci USA. 1979;76:1358–1362. doi: 10.1073/pnas.76.3.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hei T K, Waldren C A, Hall E J. Radiat Res. 1988;115:281–291. [PubMed] [Google Scholar]
- 21.Hei T K, Wu L J, Liu S X, Vannais D, Waldren C A. Proc Natl Acad Sci USA. 1997;94:3765–3770. doi: 10.1073/pnas.94.8.3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Waldren C A, Correll L, Sognier M A, Puck T T. Proc Natl Acad Sci USA. 1986;83:4839–4843. doi: 10.1073/pnas.83.13.4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hei T K, Piao C Q, He Z Y, Vannais D, Waldren C A. Cancer Res. 1992;52:6305–6309. [PubMed] [Google Scholar]
- 24.Watanabe M, Suzuki M, Suzuki K, Hayakawa Y, Miyazaki T. Radiat Res. 1990;124:73–78. [PubMed] [Google Scholar]
- 25.Littlefield L G, Joiner E E, Colyer S P, Sayer A M, Frome E L. Int J Radiat Biol. 1988;53:875–890. doi: 10.1080/09553008814551241. [DOI] [PubMed] [Google Scholar]
- 26.Pelletier J, Bruening W, Kashtan C E, Mauer S M, Manivel J C, Striegel J E, Houghton D C, Junien C, Habib R, Fouser L, et al. Cell. 1991;67:437–447. doi: 10.1016/0092-8674(91)90194-4. [DOI] [PubMed] [Google Scholar]
- 27.Vasicek T J, McDevitt B E, Freeman M W, Fennick B J, Hendy O N, Potts J T, Jr, Rich A, Kronenburg H M. Proc Natl Acad Sci USA. 1983;80:2127–2131. doi: 10.1073/pnas.80.8.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karathanasis S K, Zannis V I, Breslow J L. Proc Natl Acad Sci USA. 1983;80:6147–6151. doi: 10.1073/pnas.80.20.6147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu L X, Waldren C A, Vannais D, Hei T K. Radiat Res. 1996;145:251–259. [PubMed] [Google Scholar]
- 30.McGuinness S M, Shibuya S M, Ueno A M, Vannais D, Waldren C A. Radiat Res. 1995;142:247–255. [PubMed] [Google Scholar]
- 31.Wang T S, Huang H. Mutagenesis. 1994;9:253–257. doi: 10.1093/mutage/9.3.253. [DOI] [PubMed] [Google Scholar]
- 32.Zhao C Q, Young M R, Diwan B A, Coogan T P, Waalkes M P. Proc Natl Acad Sci USA. 1997;94:10907–10912. doi: 10.1073/pnas.94.20.10907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lynn S, Lai H T, Gurr G R, Jan K Y. Mutagenesis. 1997;12:353–358. doi: 10.1093/mutage/12.5.353. [DOI] [PubMed] [Google Scholar]
- 34.Call K M, Ito C Y, Lindberg C, Memisoglu A, Petrou C, Glaser T, Jones C, Housman D E. Somat Cell Mol Genet. 1992;18:463–475. doi: 10.1007/BF01233086. [DOI] [PubMed] [Google Scholar]
- 35.Evans H H. Radiat Res. 1994;137:131–144. [PubMed] [Google Scholar]
- 36.Baker R M, Brunette D M, Manlovitz R, Thompson L H, Whitmore G, Simmovitch L, Till J E. Cell. 1974;1:9–21. [Google Scholar]
- 37.Moore M M, Harrington-Brock K, Doerr C L. Mutat Res. 1997;386:279–290. doi: 10.1016/s1383-5742(97)00003-3. [DOI] [PubMed] [Google Scholar]
- 38.Nordenson I, Beckman L. Hum Hered. 1991;41:71–73. doi: 10.1159/000153979. [DOI] [PubMed] [Google Scholar]
- 39.Barchowsky A, Dudek E J, Treadwell M D, Wetterhahn K E. Free Radical Biol Med. 1996;21:783–790. doi: 10.1016/0891-5849(96)00174-8. [DOI] [PubMed] [Google Scholar]
- 40.Moody C S, Hassan H M. Proc Natl Acad Sci USA. 1982;79:2855–2858. doi: 10.1073/pnas.79.9.2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oller A R, Thilly W G. J Mol Biol. 1992;228:813–826. doi: 10.1016/0022-2836(92)90866-i. [DOI] [PubMed] [Google Scholar]
- 42.Hsie A W, Xu W, Yu Y, Sognier M A, Hrelia P. Tertaog Carcinog Mutagen. 1990;10:115–124. doi: 10.1002/tcm.1770100207. [DOI] [PubMed] [Google Scholar]
- 43.Kruszewski M, Green M H L, Lowe J E, Szumiel I. Mutat Res. 1994;308:233–241. doi: 10.1016/0027-5107(94)90158-9. [DOI] [PubMed] [Google Scholar]