Abstract
We have developed a new method for identifying specific single- or double-stranded DNA sequences called nicking endonuclease signal amplification (NESA). A probe and target DNA anneal to create a restriction site that is recognized by a strand-specific endonuclease that cleaves the probe into two pieces leaving the target DNA intact. The target DNA can then act as a template for fresh probe and the process of hybridization, cleavage and dissociation repeats. Laser-induced fluorescence coupled with capillary electrophoresis was used to measure the probe cleavage products. The reaction is rapid; full cleavage of probe occurs within one minute under ideal conditions. The reaction is specific since it requires complete complementarity between the oligonucleotide and the template at the restriction site and sufficient complementarity overall to allow hybridization. We show that both Bacillus subtilis and B. anthracis genomic DNA can be detected and specifically differentiated from DNA of other Bacillus species. When combined with multiple displacement amplification, detection of a single copy target from less than 30 cfu is possible. This method should be applicable whenever there is a requirement to detect a specific DNA sequence. Other applications include SNP analysis and genotyping. The reaction is inherently simple to multiplex and is amenable to automation.
INTRODUCTION
Hybridization provides the basis for specific nucleotide sequence detection in a number of techniques commonly used in molecular biology. These include microarrays, polymerase chain reaction (PCR) (1,2), Southern blotting (3), rolling circle amplification (4) and many others (5). All hybridization-based methods require small oligonucleotides, primers or probes, to recognize specific sequences in target DNA and specifically hybridize to these target regions as part of the detection process. Discrimination of such primers or probes between identical and related DNA sequences requires precise control of oligonucleotide Tm, hybridization temperature and salt concentration.
Hybridization-based methods are commonly used to identify organisms present in environmental or biological samples. All have both strengths and weaknesses. Microarrays, for example, are very specific because multiple probes can be used in parallel to interrogate different regions of a genome (6,7). However, microarrays are not the most sensitive technique with reported limits of detection of around 1000 to 2000 cells (7,8). PCR on the other hand is both specific and highly sensitive (LODs of 10 or fewer organisms) but it is inhibited by contaminants commonly found in both environmental and biological samples. As a result, stringent isolation and purification pre-processing procedures are required to avoid false negatives (7,9,10). We have developed a hybridization-based nucleic acid detection method that is specific and, when combined with multiple displacement amplification, is sensitive and tolerant to contaminants commonly found in biological samples (10).
Restriction enzymes classically recognize a double-stranded DNA-binding site, and cleave each strand of the DNA using two independent catalytic cleavage centers (11). In contrast, nicking endonucleases such as Nt.BstNBI (12), only cut one strand. Nt.BstNBI is a naturally occurring nicking endonuclease that only cleaves one strand due to its inability to form dimers. New England BioLabs engineered the nicking endonuclease Nt.AlwI by creating a chimeric protein, which consists of the DNA-binding domain and catalytic center of AlwI fused to the defective dimerization domain of Nt.BstNBI (13). We have used the single-strand cleavage activities of Nt.Alw1 to develop a sensitive method for detecting the presence of unique DNA sequences that contain a nicking endonuclease recognition site. The presence of a restriction site within the probe increases specificity since hybridization alone is not enough for enzyme recognition. Instead, an exact sequence match is usually required [see ref. (14) and New England BioLabs catalog for details on DNA binding and the effects of buffer solutions on DNA cleavage specificity]. In this system, the probe binds to its target and is cleaved into two pieces that then dissociate from the target. More full-length probe then binds to the target, and the process continues until either all the probe oligonucleotides are cleaved or the enzyme is no longer active. We call this reaction nicking endonuclease signal amplification (NESA) since multiple probes are cleaved per target DNA molecule.
MATERIALS AND METHODS
Oligonucleotides, genomic DNA and nicking enzyme
All oligonucleotides were obtained from Integrated DNA Technologies (Coralville, IA, USA). Bsub 3 (5′ 6-FAM™/TTT GGA TCG TTT CAA AGA GAG), Bsub 6 (5′ 6-FAM™/CCG GAT CTG AGG TAA CGA TGT) and BA1 (5′ 6-FAM™/AGG ATC GAA TAA GAG GTC CTT CAT) are 5′-fluorescently labeled oligonucleotide probes that were purified by ion exchange HPLC. They are identical to bases 1335052–1335072 and 431471–431491 of the Bacillus subtilis strain 168 genome (15) and bases 4610697–4610674 of the B. anthracis Ames strain respectively. Bsub 3c and Bsub 6c are the non-fluorescently labeled complementary oligonucleotides of Bsub 3 (CTC TCT TTG AAA CGA TCC AAA) and Bsub 6 (ACA TCG TTA CCT CAG ATC CGG) and were purified by standard desalting methods. Five oligonucleotide size standards (5, 17, 20, 30 and 50 bases) were synthesized with hexachlorofluorescein (HEX™) on their 5′ ends. Genomic DNA for B. subtilis, B. cereus, B. thuringiensis and B. anthracis was obtained from Dr Kevin O’Connell of the U.S. Army Edgewood Chemical Biological Center, Aberdeen Proving Ground, MD.
The nicking enzyme, Nt.Alw1, was obtained from New England BioLabs (Ipswich, MA, USA). Nt.Alw1 (10 units = 0.09 pmol) has a specific activity of 1.8 × 106 U/mg (Richard Grandoni, personal communication) and a reported turnover rate of 120 cleavage events per hour per pmol (13).
Whole genome amplification
Genomic DNA was amplified by multiple displacement amplification (MDA) (16). MDA uses Phi29 polymerase and random hexamers to uniformly amplify the entire genome of an organism with little to no amplification bias (17). MDA was performed using the REPLI-g kit from Qiagen. Ten nanograms of genomic DNA from various Bacillus species were amplified according to the manufacturer's instructions. Amplified samples were stored at −20°C. MDA DNA was quantified using the PicoGreen assay using the manufacturer's protocol (Invitrogen), and specificity of amplification by PCR analysis. The following primers were used: B. subtilis 5′-TGATCTTAGTTGCCAGCATTCAGTT, 5′-TCTGTCCATTGTAGCACGTGTGTAG; B. anthracis, 5′-GAGAAAGATGAGTAAAAAACAACAA, 5′-CATTTGTGCTTTGAATGCTAG. Bacillus subtilis genomic and MDA DNA were compared using qPCR. qPCR was performed on a BioRad iCycler iQ System using the Sybr Green assay with the following conditions: 95°C for 3 min, 95°C for 30 s, 55°C for 30 s, 72°C for 30 s with all but the 3′ at 95°C step repeated 40 times. The reactions contained 25 pmol of the primers (above), Sybr Green Supermix (BioRad) and target DNA (1 pg to 10 ng) in a total volume of 50 µl. MDA DNA had a slightly higher allele frequency than genomic DNA (<1.4-fold). This allele bias is within the range reported previously (17).
Culture analysis of B. subtilis cells
A B. subtilis culture was plated overnight on LB-agar. A single colony was isolated and grown for 4 h in 15 ml of LB broth shaken at 230 r.p.m. The culture was then diluted 1:100 with LB broth. This 1:100 dilution was further diluted into five consecutive 1:10 serial dilutions in LB broth. From each of the six dilutions, 1 µl of the dilution was mixed into 25 µl of LB broth and plated onto an LB-agar plate for incubation overnight at 37°C. Colonies were counted for each dilution the following morning. An additional 1 µl of the dilutions were used as template for an MDA reaction, for a total of six MDA reactions. The MDA reactions were performed using the REPLI-g kit from Qiagen according to the manufacturer's instructions except that 2.0 μl instead of 2.5 μl PBS were added to the cellular material to account for the additional volume from the B. subtilis culture. Amplified samples were stored at −20°C.
Nicking endonuclease amplification reaction
Each reaction contained 1× NEBuffer 2 (New England BioLabs), 1 pmol fluorescently labeled probe, 10 U (0.09 pmol) Nt.AlwI and a specified amount of target DNA, all in a total volume of 10 µl. The amount of target DNA, the length of reaction and the temperature used varied from experiment to experiment. All reactions were incubated in a PTC–200 DNA Engine thermal cycler from Bio-Rad Laboratories (Hercules, CA, USA) using the following conditions. Buffer, probe and target were added to a thin-walled PCR tube or to a PCR microplate. Samples were then incubated at 95°C for 10 min to denature the target, followed by equilibration at the specified reaction temperature for 5 min prior to addition of the enzyme. After a specified period of time, the reactions were heat-killed at 80°C for 20 min and stored at 4°C until analysis by capillary electrophoresis. All reactions were performed in triplicate.
Capillary gel electrophoresis
Samples were analyzed using an Applied Biosystems 3130xl Genetic Analyzer; electrokinetic loading was used in all cases. The distance from the loading point to the detector was 35 cm. POP 6 polymer from Applied Biosystems was used with an injection voltage of 1.2 kV and a loading time of 18 s. Run voltage was 15 kV for 10 min; oven temperature was set at 60°C. Prior to loading on the ABI3130xl, the reactions were diluted 100-fold (20-fold with water and then 5-fold with formamide). Final loading volume was 10 µl. A set of Hex™-labeled standards was run with each sample to aid in peak identification, which was performed using GeneMapper software from Applied Biosystems.
RESULTS
Nicking endonuclease signal amplification
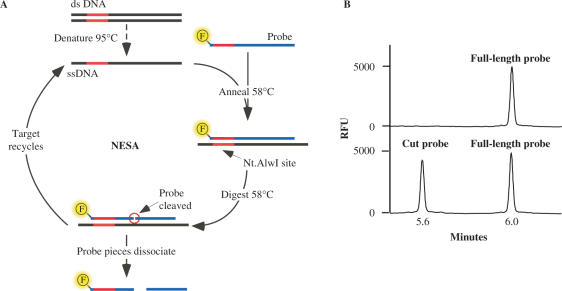
NESA is a sensitive method for identifying specific single- or double-stranded DNA sequences. The method involves the hybridization of a complementary oligonucleotide probe to the target DNA (Figure 1). Target DNA is first denatured by heating at 95°C for 10 min in the presence of a molar excess of an oligonucleotide probe. The target DNA can be any DNA that is, or can be made, single-stranded. We have used oligonucleotides, PCR-amplified DNA, and whole genome amplified (WGA) genomic DNA (amplified by multiple displacement amplification). The probe is an oligonucleotide that has a fluor on the 5′ or 3′ end or internally. The probe is complementary to one strand of the target DNA and when annealed, the oligonucleotide probe and target create a recognition site for a strand-specific nicking endonuclease that cleaves the oligonucleotide probe into two pieces, but leaves the target intact. Once the full-length probe is cleaved in two, the affinity of the probe fragments for target DNA is reduced and they dissociate leaving the target free to again form a complex with full-length oligonucleotide probe.
Figure 1.
Nicking endonuclease signal amplification. (A) Double-stranded target DNA containing an Nt.Alw I site is first denatured at 95°C and then annealed to a fluorescently labeled oligonucleotide probe at 58°C. Theoretically, the fluorescent label can be anywhere in the probe as long as it does not interfere with Nt.AlwI cleavage; we have used it successfully at either the 5′ or 3′ positions. Nt.Alw I cleaves the probe but not the target DNA and the two probe pieces spontaneously dissociate from the target. New full-length probe anneals to the target and the reaction is repeated. (B) Analysis of NESA using capillary electrophoresis. The top trace shows only full-length probe, indicating no cleavage activity occurred while the bottom trace shows an example of positive cleavage activity; the two peaks correspond to cut probe and full-length probe.
The reaction contains a molar excess of oligonucleotide probe therefore hybridization, cleavage and dissociation can occur many times. NESA is only limited, theoretically, by the availability of oligonucleotide probe or the stability of the enzyme. The reaction is highly specific since it requires complete complementarity between the oligonucleotide probe and the target at the restriction site and sufficient complementarity to allow hybridization outside of the enzyme recognition site. Hybridization temperatures can be adjusted to allow increased or decreased specificity; sequences containing just one mismatch (e.g. single nucleotide polymorphisms) are easily distinguished if desired.
NESA cleaves the probe into two pieces, one of which is fluorescently labeled. Since the resulting pieces are smaller than the full-length probe, the rate of the reaction can be measured by any method that can both detect fluorescent molecules and distinguish such oligonucleotides by size. In this study, we used capillary electrophoresis (CE) and laser induced fluorescence (i.e. an automated DNA sequencer) to measure these parameters at specific endpoints (Figure 1B). Measurement by CE is fast, sensitive and amenable to high throughput.
Temperature range of NESA
In general, sequence-specific DNA detection using hybridization reactions requires a hybridization temperature that allows binding of the probe to its cognate sequence but not to related sequences. Specificity improves as the length of the probe increases so long as the hybridization temperature can also be increased. In NESA, probe hybridization is immediately followed by nicking endonuclease cleavage of the probe and dissociation of the two smaller DNA pieces. Thus in our system there are two additional constraints: the hybridization temperature needs to be compatible with the thermostability of the nicking endonuclease and the temperature has to be high enough to allow rapid dissociation of the cleaved probe.
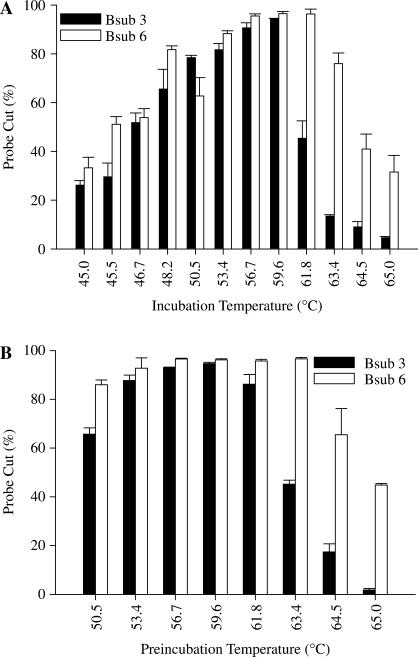
Nt.AlwI has a recommended incubation temperature of 37°C based on the standard nicking endonuclease reaction in which supercoiled, circular DNA is nicked. To determine whether higher temperatures could be used for NESA, we performed a series of standard reactions using incubation temperatures up to 65°C (Figure 2A). We used two 21-mer B. subtilis probes, Bsub 3 and Bsub 6, that have a standard Tm of 50.6 and 56.3°C, respectively (Integrated DNA Technologies Inc). Both probes had a relatively small amount of activity at 45°C, a broad optimum temperature range between 50 and 60°C and a precipitous drop in activity above 60°C. The Bsub 6 probe had slightly higher activity and retained activity longer than Bsub 3.
Figure 2.
Effects of temperature on NESA. Reactions were set up as detailed using the standard NESA procedure described in Methods using the indicated fluorescent probes and 10 fmol of each complement oligonucleotide. Reactions were then analyzed using capillary electrophoresis. (A) Reactions were incubated for 1 h at the indicated temperatures. (B) Buffer, probe and enzyme were incubated together at the indicated temperatures for 15 min. Reactions were then equilibrated at 58°C, complement added, and incubation continued for 1 h at 58°C.
The precipitous drop in activity above 60°C was very suggestive of a protein denaturation event. To determine if this was the case, we pre-incubated reactions without target complement DNA at set temperatures for 15 min, equilibrated the reactions at 58°C and then added complement to start the reaction (Figure 2B). Pretreatment up to ∼60°C had little effect on enzymatic activity but higher temperatures led to a similar precipitous drop in activity as seen in Figure 2A, suggesting that indeed the limitation in incubation temperature is due to enzyme denaturation. There does appear to be a small probe-specific effect since reactions containing Bsub 6 retained activity longer. Based on these results, we decided to use a standard incubation temperature of 58°C. Successive experiments showed that Nt.Alw1 retained full activity for at least 1 h at 58°C (data not shown).
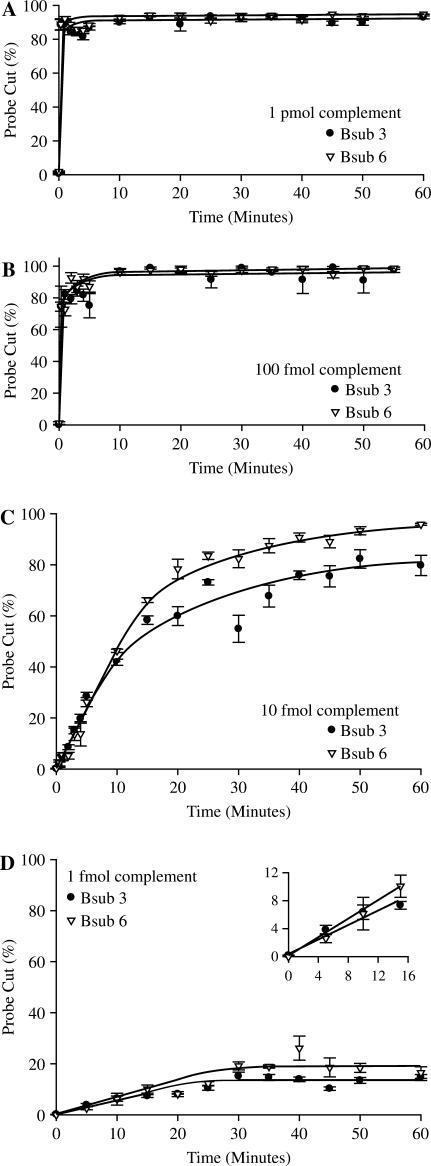
Reaction rate and target concentration
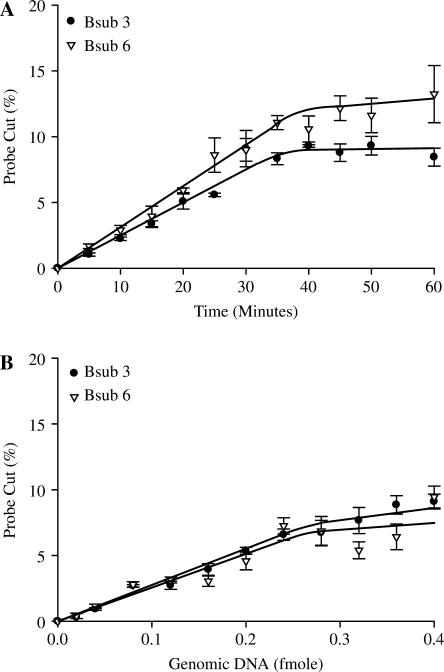
NESA is extremely rapid. Using equimolar concentrations of probe and complement target (1 pmol) and 10 U of enzyme, the reaction is almost instantaneous with over 90% cleavage of probe occurring within 1 min (Figure 3A). By decreasing the target concentration 10-fold to 100 fmol (Figure 3B), the reaction slows down somewhat but still shows more than 70% cleavage within 1 min and the reaction has gone to completion by 10 min. As target is reduced even further (Figure 3C and D), the reaction rate is further decreased and at 1 fmol the reaction plateaus at ∼15% cleavage after 30 min. As evident in the inset of Figure 3D, there is an early linear response that then plateaus. This plateau is consistent with a decline in reaction rate seen when reaction components become limited. One femtomole is the approximate limit of detection of oligonucleotide targets using our standard reaction and detection technique since at 0.1 fmol we observe very little activity (data not shown).
Figure 3.
Sensitivity using oligo targets. NESA reactions were set up as detailed using the standard procedure described in Methods section using the indicated fluorescent probes and the indicated amount of each complement oligonucleotide. Reactions were allowed to run for the indicated time at 58°C and then analyzed using capillary electrophoresis.
Based on the results with the oligonucleotide probes and their complements, which we thought would be ideal substrates for NESA, we expected the assay to be less sensitive with more complex DNA. To our surprise this did not happen. We found that the limits of detection are lower with WGA genomic DNA. We can reliably detect less than 0.1 fmol of WGA genomic DNA (Figure 4). However, we never achieved the high cleavage rates seen with the higher amounts of complement (>1 fmol) when using WGA genomic DNA. Maximum cleavage was ∼15% using 0.4 fmol of genomic target (1 µg). This is more than sufficient for adequate identification since cleavage levels of as little as 1% are well over background.
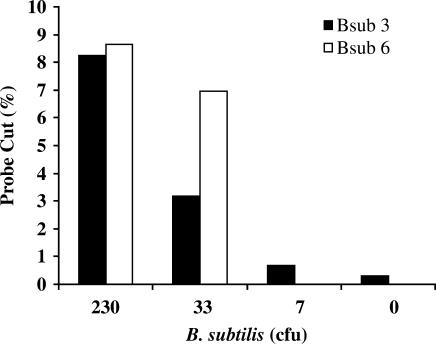
One of the possible uses of NESA is to detect and identify bacteria present in a crude sample. To determine the limits of detection of NESA in such an assay, we used serial dilutions of a log phase culture of B. subtilis. One aliquot of each dilution was plated on LB agar plates to determine the number of colony forming units (cfu) present, and the other aliquot underwent MDA, NESA and CE detection without any prior sample clean up. As shown in Figure 5, we can detect a single copy target from less than 30 cfu from unpurified sample.
Figure 5.
Limit of detection using bacterial cells. A log phase culture of B. subtilis was serially diluted and 1 µl of each dilution was used to determine colony forming units by growth overnight on LB plates. Another 1 µl aliquot from each of these dilutions was amplified directly with multiple displacement amplification, without sample clean up, then underwent NESA analysis with both Bsub 3 and Bsub 6 probes.
Specificity of NESA
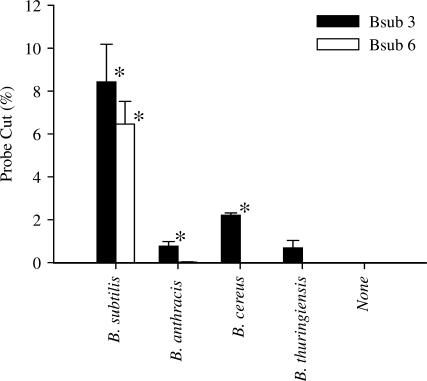
The genomic targets of Bsub 3 and Bsub 6 were chosen with little regard to specificity, although both probes represent single copy regions of the genome as their exact sequences are not present elsewhere in the genome. To determine the specificity of these probes, WGA DNA from a panel of Bacillus species was screened using our standard NESA (Figure 6). Bsub 6 recognizes B. subtilis with no significant activity against B. anthracis, B. cereus and B. thuringiensis. Bsub 3, in contrast, recognizes B. subtilis, B. anthracis and B. cereus. To show that it is possible to generate other species-specific probes, we synthesized a probe against B. anthracis (BA1, Methods section) and tested it against closely and distantly related organisms. BA1 recognized B. anthracis but had no activity at all against B. cereus, B. subtilis, B. thuringiensis, mouse and human DNA (data not shown).
Figure 6.
Probe specificity. Standard NESA reactions were performed using 0.2 fmol of genomic DNA from various Bacillus species. No cut probe was detected in the absence of added DNA or using the Bsub 6 probe with B. anthracis, B. cereus or B thuringiensis genomic DNA. *, significantly different (t-test) from the no added DNA control (P < 0.005).
Multiplex assays
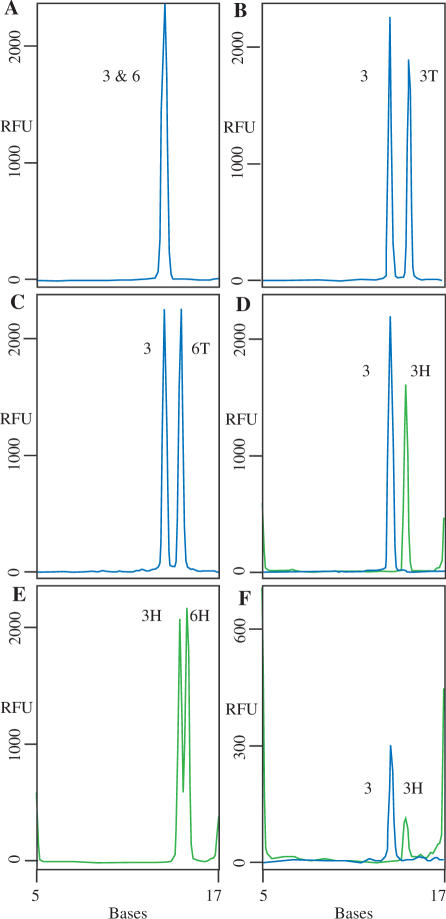
Theoretically, the NESA reaction can be multiplexed based on oligonucleotide size and by using oligonucleotides labeled with different fluors. In Figure 7A, we performed a multiplex assay using both Bsub 3 and Bsub 6 (FAM-labeled) probes and their complements. When cleaved in NESA, Bsub 3 and Bsub 6 migrated indistinguishably from each other on CE even though they are 12 and 11 bases long respectively. In order to increase the separation of the two probes, two new FAM-labeled probes were synthesized that had 2 additional bases (Ts) on the 5′ end. These probes are clearly distinguishable from the original Bsub 3 and Bsub 6 probes (Figure 7B and C). We demonstrated the ability to multiplex using probes labeled with different fluors by synthesizing an additional two probes, HEX-labeled Bsub 3 and HEX-labeled Bsub 6 (Figure 7D–F). We expected that the FAM- and HEX-labeled Bsub 3 probes would migrate at the same position and that they could be distinguished by their spectral characteristics, a routine function of the ABI3130 that we use for capillary electrophoresis. However, in this experiment, the use of alternate dyes in itself altered the mobility of the fragments so that the cleaved fragments of FAM-Bsub 3 and HEX-Bsub 3 could be easily distinguished both with oligonucleotide complements (Figure 7D) and with B. subtilis MDA DNA (Figure 7F).
Figure 7.
Multiplex assays. Standard NESA reactions were set up using either oligonucleotide complements (A–E) or 500 ng B. subtilis MDA DNA. Peaks labeled 3 and 6 are derived from FAM-labeled probes Bsub 3 and Bsub 6, respectively (blue lines). Peaks labeled 3T and 6T are derived from FAM-labeled Bsub 3 and Bsub 6 probes with two additional nucleotides (T2) at the 5′ end. Peaks labeled 3H and 6H are HEX-labeled Bsub 3 and Bsub 6 probes, respectively (green lines). The positions of the 5 and 17 base hex-labeled standards are shown. The cleaved probes migrate at positions that cannot be determined solely by their length. The apparent sizes of the cleaved probes are: FAM-Bsub 3, 13.4 bases; FAM-Bsub 6, 13.6 bases; FAM-Bsub 3T2, 14.6 bases; FAM-Bsub 6T2, 14.4 bases; HEX-Bsub 3 bases, 14.5; HEX-BSub 6, 15.1 bases.
DISCUSSION
The ability to detect specific DNA sequences is a common requirement of many techniques in molecular biology. A method of identifying specific DNA sequences that relies on the single-strand specific nucleolytic activity of a nicking endonuclease has been developed. NESA is rapid and sensitive, detecting a single copy target from less than 30 cfu of unpurified cellular material in less than 1 h. We have also demonstrated that the technique can be used to develop species-specific probes capable of discriminating between closely related organisms.
NESA is extremely rapid, cleaving over 90% of the probe in lesser than a minute with 10-fold less enzyme than substrate (probe). Nt.AlwI has been reported to have a turnover rate of approximately two cleavage events per minute in an experiment that was performed at the recommended incubation temperature of 37°C and on a substrate of supercoiled, circular DNA (13). Under our conditions, an incubation temperature of 58°C with a linear oligonucleotide substrate, we have achieved turnover rates upwards of 10 cleavage events per minute (Figure 3A). Preliminary experiments with 100-fold lesser enzyme than substrate indicate turnover rates as high as 16 cleavage events per minute (data not shown). The increased turnover rate in NESA compared to the supercoil relaxation assay could be simply due to the increased temperature (58°C for NESA versus 37°C) or it could be due to the destruction of the substrate in NESA (at 58°C the products dissociate) but not in the supercoil relaxation assay.
NESA requires a single-strand DNA target and we have shown that robust signals can be generated from both short oligonucleotides and longer, more complex genomic DNA. In reactions containing more than 10 fmol of oligonucleotide target DNA, we routinely obtain near complete cleavage of the probe. At lower oligonucleotide target levels or with genomic targets, we get lower levels of probe cleavage. Even so, we reliably achieve 5–15% cleavage of the input probe using WGA genomic DNA as target. Decreased rates of probe cleavage occur with low levels of both genomic and oligonucleotide targets (Figures 3 and 4), therefore the phenomenon appears not to be specific to the type of target DNA. We know that this effect is not due to loss of enzyme activity since Nt.AlwI preincubated at 58°C for 1 h is as active as fresh enzyme. Similarly, it is not due to insufficient enzyme since the addition of more enzyme halfway through the reaction does not increase the signal. In the case of WGA genomic DNA, there is an indication that the lower rates may be due, at least partially, to probe access issues since sonication of the genomic DNA can increase the signal up to 25% (data not shown).
Figure 4.
Sensitivity using genomic targets. Standard NESA reactions were set up using B. subtilis WGA DNA. (A) Reactions contained 0.2 fmol WGA DNA and were incubated at 58°C for the indicated times. (B) NESA reactions containing the indicated amounts of MDA DNA were run for 60 min at 58°C.
The 10-fold increase in sensitivity with genomic DNA compared to oligonucleotide target was unexpected since genomic DNA is not only more complex but also contains many Nt.AlwI sites outside of the target sequence that, if re-annealed, could compete for enzyme. It does not appear to be due to the presence of multiple targets within the genomic DNA because, based upon sequence analysis, the probes were designed to single copy regions of the B. subtilis genome. One possibility is that the longer DNA allows the nicking endonuclease to search for recognition sites in 1D space (14), rather than by 3D diffusion as with short oligos. Another possibility is that the products of the reaction, cut probe pieces, bind transiently to the oligonucleotide target and thus, compete with full-length probe for the target sites. This should be less of a problem with complex DNA since there are plenty of sites for the probe pieces to transiently bind other than the actual target sites. Therefore, the target sites remain available for binding with the full-length probe. A final possibility is that the single-stranded regions on either side of probe hybridized to genomic DNA increase cleavage efficiency. This seems unlikely, however, as single-stranded regions next to a restriction enzyme binding site have not been shown to contribute to cleavage efficiency. Indeed, we have found that even the length of a double stranded 5′ extension has little effect on the cleavage reaction. For instance, Bsub 3 has 3 bp 5′ of the restriction enzyme recognition site while Bsub 6 has a 2 bp overhang, yet they have similar sensitivities on both genomic and oligo targets. In addition, we have found that probes with only 1 bp 5′ of the recognition site work well.
NESA can be used as a method to detect specific organisms based on their DNA sequence. One of the common problems with current detection methods is the requirement to purify the DNA away from contaminants prior to analysis. When combined with multiple displacement amplification (MDA), NESA can be used to detect specific sequences from crudely prepared samples (Figure 5). MDA has the ability to amplify DNA from an unpurified sample because it is tolerant of the contaminants commonly found in biological or environmental samples (10,17). Preliminary studies indicate that MDA can amplify DNA from unpurified environmental samples in cases where real-time PCR fails (data not shown).
While NESA already exhibits excellent sensitivity, detecting a single DNA target in less than 30 cfu (Figure 5), further increases in sensitivity are possible by optimization of the capillary electrophoresis step. The CE step currently requires a 100-fold dilution of the reaction. Therefore, of the 1 pmol of probe that was in the reaction, only 10 fmol is interrogated by the CE system. If the reaction can be modified such that the 1:100 dilution is not required, a 100-fold increase in sensitivity can be gained. The dilution step serves two purposes. First it decreases the ionic strength of the buffer allowing more efficient electrokinetic injection. Second, it reduces the level of fluorescent oligo that would otherwise overload the capillary. In reactions where increased sensitivity is required, most of the fluorescent signal comes from uncut probe. In preliminary experiments, we have had some success in removing full-length probe by incorporating biotin into the fluorescent probe and then using streptavidin conjugated to magnetic beads to remove full-length probe following NESA.
The specificity of NESA is in part due to the requirement of the target sequence to contain a nicking endonuclease target site. We have found that with Nt.AlwI probes a mismatch in any one of the 5 bp that constitute the recognition sequence completely inhibits cleavage (unpublished data). However, we do not know if impurities can give rise to star activity (18). The ideal length of the probe is not known, but presumably it is partially a function of the Tm of the full-length probe and the Tm of the two products. That is, the Tm differential has to be sufficient to allow initial hybridization of the probe followed by subsequent dissociation of both probe pieces. The probe also requires a certain length to retain its uniqueness in terms of base-pair composition, and thus its specificity for the sequence to which it was designed. Furthermore, the thermostability of the nicking enzyme must be taken into account because the enzyme becomes denatured at temperatures upwards of 60°C. Therefore, probes cannot have a Tm much greater than 60°C because specificity would have to be forfeited in order to maintain enzyme function. We analyzed two probes at an incubation temperature of 58°C, Bsub 3 and Bsub 6. Bsub 6 had a Tm of 56.3°C and was specific for B. subtilis while Bsub 3 had a lower Tm of 50.6°C and was more promiscuous detecting both B. subtilis, B. anthracis and B. cereus. Clearly, specificity is not merely a function of probe Tm as illustrated by these two examples. Instead, it is likely a combination of parameters, most notably Tm and sequence composition, that influence specificity.
NESA is also highly amenable to multiplexing since multiple sequences can be detected in a single reaction. Multiplexing by both probe size and fluorescent color is possible (Figure 7). For size discrimination, the position of the restriction site within the probe can be varied (Figure 7) or the fluorescent label can be placed at either the 5′ or 3′ end (data not shown). For color discrimination, we have shown that both FAM- and HEX-labeled probes can be used. Theoretically any fluorescent dye can be used that can be used to modify the oligonucleotide and that can be discriminated from the other dyes used in the reaction. This of course depends on the spectral characteristics of the dye and the capabilities of the capillary electrophoresis instrument used. We have found that the length of the probes is not a good predictor of their migration on CE; probes of the same length can separate while probes of different lengths can migrate together. In addition the fluor used can alter the migration pattern.
While we focused on Nt.Alw1, there are a number of other naturally occurring or designed nicking endonucleases that have been described (19,20 and references within). However, only a few are available commercially, mainly from New England BioLabs (19). We have successfully tested six enzymes [Nb.BbvCI (21), Nb.BsmI, Nb.BsrDI, Nt.AlwI (13), Nt.BbvCI (21), Nt.BstNBI (12)] in NESA but we have yet to perform a careful analysis of their relative effectiveness. Nb.BsmI and Nb.BsrDI are particularly interesting in that they are active up to at least 65°C and so should allow the development of longer, more specific probes. Nb.BbvCI and Nt.BbvCI recognize the same 7 bp DNA sequence but cleave opposite strands. The 7-bp recognition site increases specificity of the probes but also reduces the number of regions of DNA that can be targeted (Nt.AlwI recognizes 5 bp).
NESA, although designed to detect DNA sequences, theoretically can be used to detect RNA sequences as well, as long as the RNA can be converted to cDNA. In some applications, such as rapid detection of RNA viruses, the ability to detect RNA directly would be beneficial. We have not tried to use NESA with RNA targets but there is some evidence in the literature which suggests that detection of DNA/RNA hybrids by wild type (22) or genetically modified restriction enzymes (23) may be possible.
In summary, the utility of NESA was demonstrated for detection of specific DNA sequences. Several occasions can be contemplated where NESA may represent the method of choice for detection of DNA sequences. First, NESA can be used as an assay in the detection of specific organisms. In medical diagnostics, NESA can be used as an assay for bacterial and viral agents, which affect both human and animal health. Since NESA is amenable to high-throughput automation, it can be used to combat bioterrorism in stand-alone devices that test aerosol samples for agents such as B. anthracis, the causative agent of anthrax. Of course with this assay as with all other DNA-based assays, terrorists could modify the target sequences rendering the detection system ineffective. The use of multiple probes and keeping targets secret makes this countermeasure less likely. In ecological studies, NESA would be useful in species and/or strain analysis. Genomic analysis is another area of research where it may be useful. NESA can distinguish between closely related DNA sequences and detect the presence of single nucleotide mutations/polymorphisms. In an experiment, where we introduced point mutations into oligonucleotide targets at each base-pair position, we found that a single mutation at any of the 5′ positions in the enzyme recognition site is sufficient to completely eliminate the signal (data not shown).
ACKNOWLEDGEMENTS
This work was supported by contracts W911SR–05–C–0029 and W911SR–04–C–0036 from the US Department of Defense. We thank Kevin O’Connell, Edgewood Chemical Biological Command, for his guidance and Ryan Elwell, Ryan Mahnke, Nitu Thakore, Jason Thompson, Zach Peksa, from Anteon Corporation and General Dynamics for their support. We also thank, Joel Credle, Tania Rozgaja and Lilit Vardanian for their technical help. Funding to pay the Open Access publication charges for this article was provided by grant number WP11SR-07-C-0026/DOC9 from the U.S. Army.
Conflict of interest statement. None declared.
REFERENCES
- 1.Saiki RK, Scharf S, Faloona F, Mullis KB, Horn GT, Erlich HA, Arnheim N. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985;230:1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- 2.Kubista M, Andrade JM, Bengtsson M, Forootan A, Jonak J, Lind K, Sindelka R, Sjoback R, Sjogreen B, et al. The real-time polymerase chain reaction. Mol. Aspects Med. 2006;27:95–125. doi: 10.1016/j.mam.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 3.Southern EM. Blotting at 25. Trends Biochem. Sci. 2000;25:585–588. doi: 10.1016/s0968-0004(00)01702-3. [DOI] [PubMed] [Google Scholar]
- 4.Lizardi PM, Huang X, Zhu Z, Bray-Ward P, Thomas DC, Ward DC. Mutation detection and single-molecule counting using isothermal rolling-circle amplification. Nat. Genet. 1998;19:225–232. doi: 10.1038/898. [DOI] [PubMed] [Google Scholar]
- 5.Andras SC, Power JB, Cocking EC, Davey MR. Strategies for signal amplification in nucleic acid detection. Mol. Biotechnol. 2001;19:29–44. doi: 10.1385/MB:19:1:029. [DOI] [PubMed] [Google Scholar]
- 6.Vora GJ, Meador CE, Stenger DA, Andreadis JD. Nucleic acid amplification strategies for DNA microarray-based pathogen detection. Appl. Environ. Microbiol. 2004;70:3047–3054. doi: 10.1128/AEM.70.5.3047-3054.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lim DV, Simpson JM, Kearns EA, Kramer MF. Current and developing technologies for monitoring agents of bioterrorism and biowarfare. Clin. Microbiol. Rev. 2005;18:583–607. doi: 10.1128/CMR.18.4.583-607.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burton JE, Oshota OJ, North E, Hudson MJ, Polyanskaya N, Brehm J, Lloyd G, Silman NJ. Development of a multi-pathogen oligonucleotide microarray for detection of Bacillus anthracis. Mol. Cell. Probes. 2005;19:349–357. doi: 10.1016/j.mcp.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 9.Yang S, Rothman RE. PCR-based diagnostics for infectious diseases: uses, limitations, and future applications in acute-care settings. Lancet Infect. Dis. 2004;4:337–348. doi: 10.1016/S1473-3099(04)01044-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gonzalez JM, Portillo MC, Saiz-Jimenez C. Multiple displacement amplification as a pre-polymerase chain reaction (pre-PCR) to process difficult to amplify samples and low copy number sequences from natural environments. Environ. Microbiol. 2005;7:1024–1028. doi: 10.1111/j.1462-2920.2005.00779.x. [DOI] [PubMed] [Google Scholar]
- 11.Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE – restriction enzymes and DNA methyltransferases. Nucleic Acids Res. 2005;33:D230–D232. doi: 10.1093/nar/gki029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morgan RD, Calvet C, Demeter M, Agra R, Kong H. Characterization of the specific DNA nicking activity of restriction endonuclease N.BstNBI. Biol. Chem. 2000;381:1123–1125. doi: 10.1515/BC.2000.137. [DOI] [PubMed] [Google Scholar]
- 13.Xu Y, Lunnen KD, Kong H. Engineering a nicking endonuclease N.AlwI by domain swapping. Proc. Natl Acad. Sci. USA. 2001;98:12990–12995. doi: 10.1073/pnas.241215698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pingoud A, Jeltsch A. Structure and function of type II restriction endonucleases. Nucleic Acids Res. 2001;29:3705–3727. doi: 10.1093/nar/29.18.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kunst F, Ogasawara N, Moszer I, Albertini AM, Alloni G, Azevedo V, Bertero MG, Bessieres P, Bolotin A, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 16.Dean FB, Hosono S, Fang L, Wu X, Faruqi AF, Bray-Ward P, Sun Z, Zong Q, Du Y, et al. Comprehensive human genome amplification using multiple displacement amplification. Proc. Natl Acad. Sci. USA. 2002;99:5261–5266. doi: 10.1073/pnas.082089499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hosono S, Faruqi AF, Dean FB, Du Y, Sun Z, Wu X, Du J, Kingsmore SF, Egholm M, et al. Unbiased whole-genome amplification directly from clinical samples. Genome Res. 2003;13:954–964. doi: 10.1101/gr.816903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nasri M, Thomas D. Relaxation of recognition sequence of specific endonuclease HindIII. Nucleic Acids Res. 1986;14:811–821. doi: 10.1093/nar/14.2.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chan S-H, Xu S-Y. Nicking endonucleases: the discovery and engineering of restriction enzyme variants. NEB expressions. 2006;1.2:4–5. [Google Scholar]
- 20.Samuelson JC, Zhu Z, Xu SY. The isolation of strand-specific nicking endonucleases from a randomized SapI expression library. Nucleic Acids Res. 2004;32:3661–3671. doi: 10.1093/nar/gkh674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heiter DF, Lunnen KD, Wilson GG. Site-specific DNA-nicking mutants of the heterodimeric restriction endonuclease R.BbvCI. J. Mol. Biol. 2005;348:631–640. doi: 10.1016/j.jmb.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 22.Molloy PL, Symons RH. Cleavage of DNA.RNA hybrids by type II restriction enzymes. Nucleic Acids Res. 1980;8:2939–2946. doi: 10.1093/nar/8.13.2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim YG, Shi Y, Berg JM, Chandrasegaran S. Site-specific cleavage of DNA-RNA hybrids by zinc finger/FokI cleavage domain fusions. Gene. 1997;203:43–49. doi: 10.1016/s0378-1119(97)00489-7. [DOI] [PubMed] [Google Scholar]