Abstract
The evolution of interspecies differences in morphology requires sufficient within-species variation in developmental regulatory systems on which evolutionary forces can act. Molecular analyses of naturally occurring alleles of the Arabidopsis thaliana CAULIFLOWER locus reveal considerable intraspecific diversity at this floral homeotic gene, and the McDonald–Kreitman test suggests that this gene is evolving in a nonneutral fashion, with an excess of intraspecific replacement polymorphisms. The naturally occurring molecular variation within this floral regulatory gene is associated with functionally different alleles, which can be distinguished phenotypically by their differential ability to direct floral meristem development.
Keywords: development/ecotype/flower/inflorescence/MADS-box
Morphological evolution proceeds in part by alterations in the timing and location of developmental events (1, 2). Genetic analyses have demonstrated that several classes of regulatory genes choreograph the process of morphological development (2–5), and there thus has been considerable debate as to the role regulatory gene variation plays in the evolution of novel morphological structures (1, 5, 6). It remains unclear, however, to what extent there is sufficient intraspecific variation at these developmental regulatory loci available for adaptive diversification (7) and what evolutionary forces shape this variation at the molecular level. For at least one plant species, Clarkia concinna, it has been shown that naturally occurring alleles at the bicalyx locus are responsible for the high frequency of natural floral homeotic variation in a wild population (8). Selection studies on bristle number (9) and on the ether-induced bithorax phenocopy (10) also suggest that natural diversity in regulatory loci may be associated with developmental variation in Drosophila.
The Arabidopsis thaliana gene CAULIFLOWER (CAL) belongs to the MADS-box regulatory gene family, whose members encode sequence-specific DNA-binding transcriptional activators (3, 6). This gene is one of several loci that act early in the Arabidopsis floral developmental pathway (3). In wild-type plants, CAL is expressed in flanking primordia that develop along the inflorescence axis, and this gene appears to be involved in specifying the identity of these lateral primordia as floral meristems (11). Genetic and molecular analyses reveal that the developmental activity of CAL is partially redundant to that of another floral meristem identity gene, APETALA1 (AP1) (11, 12). Phylogenetic analysis indicates that these two genes are paralogous to one another and arose as a result of a gene duplication event that may have occurred early in the evolution of the family Brassicaceae (13). Plants that are homozygous for mutant alleles at both AP1 and CAL display the cauliflower phenotype, a characteristic indeterminate proliferation of inflorescence meristems that arises from the inability to switch the identity of the lateral meristems from inflorescences to flowers. Plants that are homozygous only for mutant CAL alleles appear to produce a greater number of axillary inflorescences (12), which is consistent with an associated reduction in floral meristem specifying activity.
Molecular analysis of sequence diversity permits us to determine the levels of genetic variation at CAL, and the patterning of this diversity provides information on the evolutionary forces that shape the diversification of this regulatory locus. We demonstrate here that considerable genetic variation exists for at least one plant regulatory locus that appears to be evolving in a nonneutral fashion and that this variation is associated with differences in the ability of naturally occurring alleles to direct morphological development.
MATERIALS AND METHODS
DNA Techniques and Genetic Tests.
The A. thaliana ecotypes were obtained from single-seed propagated material provided by the Arabidopsis Biological Resource Center or the Nottingham Stock Centre. The Kent (CS6054), Bretagne (CS6615), and Firenzi (CS6555) alleles were from the population collection of P. H. Williams maintained at the Arabidopsis Biological Resource Center. The cal1–1 and cal(Ws) alleles are equivalent. Arabis lyrata seed was provided by C. H. Langley (University of California, Davis).
Miniprep DNA was isolated from young leaves as described (14). PCR was performed with 40 cycles of 1 min at 95°C, 1 min at 52°C, and 3 min at 72°C followed by 15 min at 72°C. A TAQ:Pfu polymerase mixture (Stratagene) was used to minimize nucleotide misincorporation. The CAL-specific primers AGL10.3 (for exon 2 forward) and AGL10.11 (for 3′ end reverse) were used in the PCRs. For isolating the A. lyrata CAL gene, PCR primers were designed to amplify the entire coding sequence of the gene. Amplified DNA was cloned by using the TA cloning kit (Invitrogen), and at least 5–10 independent colonies were mixed to provide plasmid DNA for sequencing. DNA sequencing was done by using an ABI377 automated sequencer (Iowa State University), and all observed singleton changes were confirmed either visually or by resequencing. The DNA sequences are available from GenBank (accession nos. AF061401 to AF061416).
CAL alleles from wild ecotypes were crossed either to ap1–1/ap1–1, cal1–1/+, or ap1–1/ap1–1,+/+ tester lines. At least 2–8 F1 plants were selfed, and 100–200 individuals from each plant were screened at the F2 for wild-type, apetala1, or cauliflower phenotypes. Plants were grown in long-day conditions at 18°C/22°C
Data Analysis.
Sequences used in this study were aligned visually. Phylogenetic analyses were conducted by using paup 3.1 (maximum parsimony) (15) and mega 1.1 (neighbor-joining) (16). For maximum parsimony, the heuristic search algorithm using the tree bisection–reconnection procedure was used, with the A. lyrata CAL sequence as the outgroup. In the neighbor-joining analysis, the Kimura-2P distance measure was used. Both analyses were done with 500 bootstrap replicates of the data. The polymorphism data were analyzed by using the sites program (17). The Tajima (18) and Fu and Li (19) tests were run without outgroup, with significance of the test statistics based on the power analysis of Simonsen et al. (20). Tests for recombination were based on the method of Hudson and Kaplan (21).
RESULTS AND DISCUSSION
Nucleotide Variation at the Arabidopsis CAL Gene.
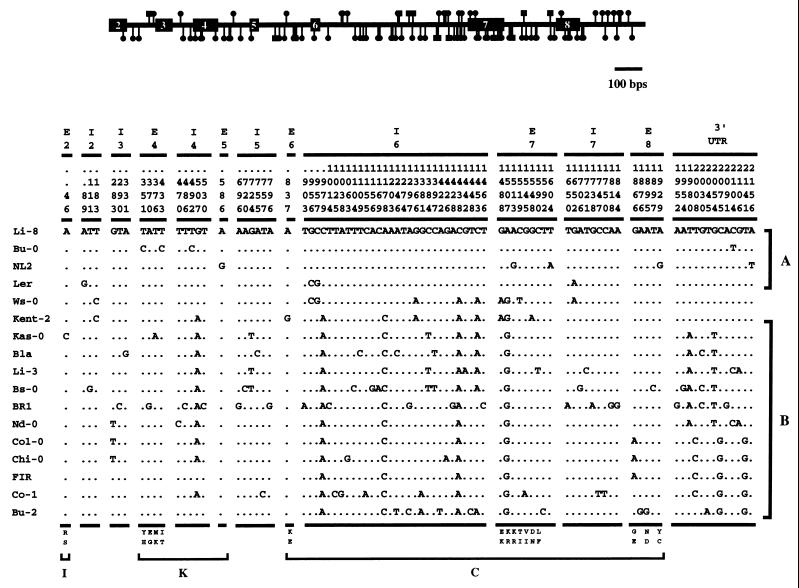
Seventeen alleles were sampled from distinct Arabidopsis ecotypes collected worldwide. Approximately 2.2 kb of this gene was sequenced for each naturally occurring allele; the sequenced region spans exons 2–8 and includes 255 bp of the 3′ flanking region of the gene (see Fig. 1). The exons sequenced encode the moderately conserved K-domain of the plant MADS proteins, believed to function as a dimerization structure, as well as the highly diverse I-linker and C-terminal regions (6).
Figure 1.
Distribution of polymorphisms along the Arabidopsis CAL gene. The circles and squares denote the approximate location of nucleotide and insertion/deletion polymorphisms, respectively. The actual nucleotide polymorphisms are tabulated and are partitioned according to intron (I) and exon (E) positions. The differentiation between class A and class B alleles are shown. The replacement polymorphisms are indicated at the bottom, with the upper row showing the encoded amino acid for the Li-8 allele. The distribution of exon polymorphisms according to the protein domains also are indicated.
The molecular analyses revealed a large amount of variation at the CAL locus (see Fig. 1). A total of 104 variant sites is present in these sampled alleles, of which 91 are nucleotide polymorphisms and 13 are insertion/deletions of 1–4 bp. Of these indels, all but one are in introns; the exception is a TTA insertion in exon 7 of the Burghaun/Rhon-2 allele that results in the addition of a leucine to the C-terminal region. A large fraction of the polymorphisms observed are singletons (68/104). The estimate of species-wide nucleotide diversity, π, for CAL was found to be 0.007, which is close to that observed for the Arabidopsis Adh gene (22) and is comparable to the mean found for a number of loci in Drosophila melanogaster (23).
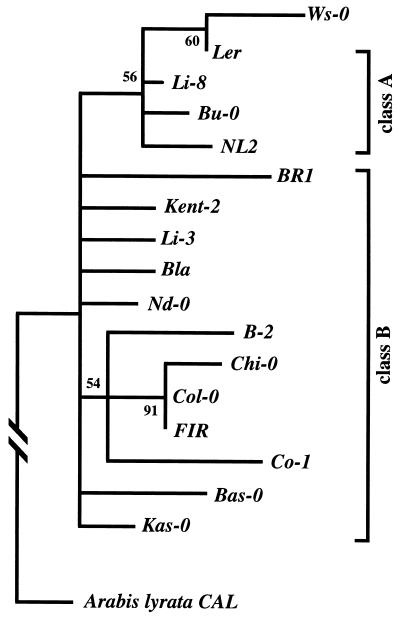
The genealogy of these naturally occurring alleles is shown in Fig. 2. The phylogeny is the result of 500 bootstrap replicates under maximum parsimony, and a tree based on neighbor-joining analysis gives the same topology. There appear to be two CAL allele classes within A. thaliana: (i) class A alleles, which include those from Landsberg erecta and three other ecotypes and (ii) class B alleles, which include alleles from the Kent-2, the Columbia, and 10 other ecotypes. The two classes are differentiated by four nucleotides and one 1-bp indel in intron 6 and exon 7 of the gene (see Fig. 1). Only one replacement polymorphism (K182 → R) distinguishes the two allele classes. There is evidence to suggest that the Ws-0 allele is a recombinant allele between a class A and B allele (see below). The phylogeny reveals no discernible geographical structuring of alleles, indicating that A. thaliana has undergone (and may continue to undergo) considerable dispersal in Europe. The CAL gene shows 3% sequence divergence from its orthologue in A. lyrata, indicating that the proliferation of CAL alleles occurred much more recently than the time of last common ancestry between these two closely related Brassicaceae species.
Figure 2.
Gene genealogy of the Arabidopsis CAL alleles based on maximum parsimony analysis. The distinctions between class A and B alleles are based on the pattern of shared polymorphisms. The numbers next to the nodes are the bootstrap values after 500 bootstrap replicates of the data; all nodes with <50% support are collapsed in the tree. Ten equally parsimonious trees (length = 528 steps) were obtained from a heuristic analysis; these trees were all congruent with the bootstrap results. The consistency index is 0.926, with the A. lyrata CAL orthologue as the outgroup. The genealogy is based on the sequence from exon 2 to the 3′ untranslated region. If only the recombinant exon 7 and flanking intron sequences are used, the Ws and Kent-2 alleles group together in the phylogeny (see text).
Nonneutral Evolution Associated with High Levels of Intraspecific Replacement Polymorphisms.
The patterning of nucleotide variation along the CAL locus suggests that this regulatory gene is evolving in a nonneutral fashion. A prediction of the equilibrium neutral theory is that the relative levels of intraspecific replacement and synonymous polymorphisms are correlated with interspecific levels of replacement and synonymous substitutions. Of the 91 polymorphic substitutions in the Arabidopsis CAL locus, 21 were found in exons. Sixteen of these sites were polymorphic replacement sites, and 5 were synonymous polymorphisms (see Table 1), suggesting an excess of intraspecific replacements within this plant species. The comparison between A. thaliana and A. lyrata revealed 13 fixed replacement and 15 fixed synonymous differences. The McDonald–Kreitman test of similarity in relative levels of replacement to synonymous changes within and between species was applied (24) and revealed a significant excess of polymorphic replacement sites within A. thaliana (P < 0.036 using Fisher’s Exact Test). This test demonstrates that the data are not in accord with the expectations based on the equilibrium neutral theory, and the null hypothesis of neutral evolution for the CAL locus can be rejected.
Table 1.
McDonald–Kreitman test for protein evolution
| Within A. thaliana polymorphisms | A. thaliana vs. A. lyrata fixed differences | |
|---|---|---|
| Replacement | 16 | 13 |
| Synonymous | 5 | 15 |
P < 0.036 using Fisher’s exact test.
Other tests for selection were applied, although these tests may be inappropriate given the high levels of inbreeding reported for this species. The Tajima (D = −1.6605, P < .075) (18) and Fu and Li (F* = −2.371, P < .057) (19) tests do indicate, however, a fairly high number of unique polymorphisms at this locus, a result that also is observed in the Adh locus (22). These results on two unlinked nuclear genes suggest that the distribution of A. thaliana worldwide occurred relatively recently. It is interesting to note that most of the replacement polymorphisms observed (13/16) are present only once in the sampled alleles.
Recombinant Alleles in the Wild.
At least two alleles, Wassileskija (from the Ukraine) and Kent-2 (from the United Kingdom), share several substitutions in and around exon 7 that differentiate them from the other alleles of the CAL gene (see Fig. 1). The Ws allele, for example, differs from the Landsberg erecta (Ler) ecotype allele at three replacement sites in this exon: a G → A transition at position 1488 (E176 → K), an A → G change at position 1507 (K182 → R), and a C → T substitution at position 1519 (T186 → I) (8). The glutamic acid at position 176 is conserved in comparisons between differing Brassicaceae CAL orthologues and even between CAL and its paralogue AP1.
Both substitutions that result in the E176 → K and K182 → R replacements, as well as 4 silent and intron polymorphisms, are shared by the Ws allele with a naturally occurring allele found in a plant from a Kent, UK population [cal(Kent-2)] (see Fig. 1). The two alleles share these polymorphisms in an ≈ 150-bp region. The sequence of the two alleles diverge outside this restricted region, where the Ws allele is similar to the Ler allele. The pattern of shared polymorphisms indicates that cal(Ws) may be a double-recombinant allele in which a restricted region of the Kent haplotype is introgressed into a Ler-like allele. Recombinant alleles also have been identified at the Adh locus (22) and the ChiA gene (25) and, together with our analysis, suggest that recombination may be an important force in shaping allelic diversity within this species.
Naturally Occurring Variation in CAL Function.
Genetic tests reveal that these different naturally occurring CAL alleles are not functionally equivalent (11, 12) and that some of these intraspecific replacement polymorphisms are associated with natural allelic variation for floral meristem specifying activity of this plant homeotic locus. The functional distinctions between different alleles at the CAL locus are distinguished readily in mutant apetala1 genetic backgrounds. Most ecotypes have CAL alleles that produce phenotypes similar to the one produced by the Landsberg erecta allele. Plants that are ap1–1/ap1–1 Cal(Ler)/Cal(Ler) give the classic apetala1 homeotic phenotype (see Fig. 3). In these lines, the loss of AP1 floral meristem identity function is compensated by the Cal(Ler) allele, resulting in the establishment of floral primordia (11). The CAL gene, however, does not have the floral organ identity function of its AP1 paralogue, and the mature flowers in these plants display homeotic modification of first and second whorl floral organs.
Figure 3.
Differences in floral phenotypes. Clockwise from upper left: (i) AP1/AP1 cal(Ws) cal(Ws), (ii) ap1–1/ap1–1 Cal(Ler)/Cal(Ler), (iii) ap1–1/ap1–1 cal(Kent-2)/cal(Kent-2), (iv) ap1–1/ap1–1 cal(Ws)/cal(Ws).
Other naturally occurring CAL alleles appear to possess a reduced ability to specify floral meristem identity in lateral primordia. The cal(Ws) allele, for example, imparts the mutant cauliflower phenotype in a homozygous ap1–1 background (see Fig. 3) (11, 12). In these plants, mutations at both AP1 and CAL result in the near-complete loss of floral meristem identity specifying activity, resulting in the proliferation of inflorescence meristems that characterize this strong phenotype. The cal(Kent-2) allele, on the other hand, does not produce a cauliflower structure. F2 progeny of Kent plants crossed in an ap1–1 background, however, produce apetala1 flowers arrayed in a denser inflorescence, and the flowers do not produce axillary floral meristems at floral bracts as in ap1–1 Cal(Ler) plants (see Fig. 3). It is unclear whether this phenotype is due to mutations in the cal(Kent-2) allele or at a closely linked locus. The CAL allele from the Nossen ecotype, however, also displays this dense inflorescence phenotype and shares with cal(Kent-2) the K182 → R replacement polymorphism at exon 7 (M. Yanofsky, personal communication). This polymorphism is one of several changes that differentiate class A and B CAL alleles (see Fig. 1). The molecular population genetic analysis, coupled with genetic investigation (11, 12), indicates that at the functional level, two distinguishable allele types are present in wild populations of A. thaliana: (i) functional Ler-like and (ii) mutant Ws/Kent-2-like alleles. The latter allele type appears to be a subset of the class B CAL alleles (see Figs. 1 and 2).
Selective Forces at the CAL Locus.
The mechanisms responsible for the high degree of intraspecific replacement polymorphism observed in the Arabidopsis CAL gene remain unclear. The data are consistent with at least three interpretations. First, the excess polymorphism observed may indicate that this locus is a pseudogene, which is possible given that both CAL and AP1 perform partially redundant floral meristem specification functions (11). There are three reasons, however, that suggest that the Arabidopsis CAL locus encodes an active gene. First, genetic tests reveal that most CAL alleles function properly in specifying floral meristem identity in Arabidopsis. Second, none of the 13 polymorphic indels observed create a frameshift or premature termination codon, which indicates that the ability of this locus to encode a functional protein may be under selection. Finally, the ratio of nonsynonymous to synonymous nucleotide substitutions at the coding region (Ka/Ks) between CAL orthologues in A. thaliana and A. lyrata is 0.271 ± 0.07. This ratio is slightly higher than the mean value of 0.146 ± 0.03 found in comparison of two other MADS-box floral homeotic genes found in these two species (see Table 2). The value of the Ka/Ks ratio for CAL, however, is still significantly <1.0, indicating that this locus remains subject to purifying selection.
Table 2.
Levels of nonsynonymous and synonymous substitutions per site between A. thaliana and A. lyrata floral homeotic genes
| Ka | Ks | Ka/Ks | |
|---|---|---|---|
| CAULIFLOWER | 0.032 ± 0.007 | 0.119 ± 0.029 | 0.271 |
| APETALA3 | 0.017 ± 0.006 | 0.113 ± 0.029 | 0.148 |
| PISTILLATA | 0.021 ± 0.007 | 0.148 ± 0.038 | 0.144 |
A second explanation is that the Arabidopsis CAL gene has experienced a recent relaxation in purifying selection. This is partly supported by the twofold increase in the Ka/Ks ratio for CAL, which is higher than the mean value for most plant nuclear genes (mean plant nuclear gene Ka/Ks = 0.19), as well as other paralogous MADS-box genes. An elevated probability of fixation of slightly deleterious replacement mutations in localized, inbred ecotype populations also may account for the overall increases in levels of within-species amino acid polymorphisms observed in A. thaliana. The partial genetic redundancy of CAL, coupled with the small effective population sizes expected from an inbreeding organism, may lead to a greater tolerance for these slightly deleterious mutations as predicted by the nearly neutral theory (26, 27). Similar high levels of intraspecific replacement polymorphisms also have been observed for mitochondrial genes, and these have been interpreted as being slightly deleterious mutations that remain at low frequency in populations (28, 29).
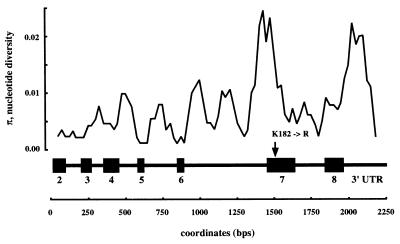
A third possibility is that the excess of within-species replacement polymorphisms may result from positive selective pressures. The number of replacement polymorphisms and the presence of two distinct haplotype classes do suggest that this locus may be under balancing selection, although the low outcrossing rates for these plants (<0.3%) would rule out overdominant selection as a mechanism for maintaining variation. There is also the possibility that selection is operating at this locus in response to the different ecological conditions that distinct ecotypes experience. Mutant CAL alleles are associated with an increase in the number of axillary inflorescences (12). An increase in the number of lateral inflorescences may result from the decrease in floral meristem-specifying activity associated with a reduction in the functional activity of Ws-like CAL proteins. Sliding window analyses indicate that a localized peak of variation associated with the K182 → R replacement polymorphism (see Fig. 4) and recent work have suggested that peaks in nucleotide diversity may arise from local selection in subdivided, inbred populations (30).
Figure 4.
Sliding window analysis of nucleotide diversity (31). The analysis is based on a 100-bp window that moves across the gene in 25-bp intervals. The exon/intron structure of the gene is indicated as a reference. The position of the K182 → R replacement polymorphism in exon 7 is indicated with an arrow.
The evolutionary pressures on regulatory genes are thought to contribute to interspecies diversification in morphology, but it has remained unclear whether sufficient variation at these developmental control genes exists on which selection can act (1, 7). The A. thaliana CAL gene provides evidence that regulatory loci that control morphological development can harbor appreciable variation at the molecular level. Moreover, the patterning of this variation suggests that regulatory loci can evolve in nonneutral fashion, resulting in the formation of functionally diverse alleles that are phenotypically distinguishable. It remains to be seen what precise evolutionary forces explain this nonneutral pattern of evolution, but our study does indicate that regulatory gene polymorphisms are present in this species and have the potential to serve as the genetic basis for evolutionary transformations in organismal development.
Acknowledgments
We thank Marty Yanofsky for providing us with the original CAULIFLOWER genomic sequence and for numerous fruitful discussions and expressions of support. We also thank Elliot Meyerowitz for providing us with the AP3 and PI genomic sequence and Katy Simonsen and members of the Purugganan laboratory for critically reading a draft of this paper, as well as the anonymous reviewers for helpful comments. Katy Simonsen also provided the statistical analysis for some the selection tests. Use of the North Carolina State University Southeastern Plant Environmental Laboratory is acknowledged gratefully. This work was supported in part by grants from the U.S. Department of Agriculture and an Alfred P. Sloan Foundation Young Investigator Award to M.D.P.
ABBREVIATIONS
- CAL
CAULIFLOWER
- AP1
APETALA1
- Ka
nonsynonymous substitutions
- Ks
synonymous substitutions, Ws, Wassileskija
Footnotes
References
- 1.Palopoli M F, Patel N H. Curr Opin Gen Dev. 1996;6:502–508. doi: 10.1016/s0959-437x(96)80074-8. [DOI] [PubMed] [Google Scholar]
- 2.Gould S J. Ontogeny and Phylogeny. Cambridge, MA: Harvard Univ. Press; 1977. [Google Scholar]
- 3.Liljegren S J, Yanofsky M F. Curr Opin Cell Biol. 1996;8:865–869. doi: 10.1016/s0955-0674(96)80089-5. [DOI] [PubMed] [Google Scholar]
- 4.Weigel D, Meyerowitz E M. Cell. 1994;78:203–209. doi: 10.1016/0092-8674(94)90291-7. [DOI] [PubMed] [Google Scholar]
- 5.Coen, E. S. & Nugent, J. M. (1994) Dev. Suppl. 107–116.
- 6.Purugganan M D, Rounsley S D, Schmidt R J, Yanofsky M F. Genetics. 1995;140:345–356. doi: 10.1093/genetics/140.1.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Richter B, Long M Y, Lewontin R C, Nitasaka E. Genetics. 1997;145:311–323. doi: 10.1093/genetics/145.2.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ford V S, Gottlieb L D. Nature (London) 1992;358:671–673. [Google Scholar]
- 9.Lai C G, Lyman R F, Long A D, Langley C H, Mackay T F C. Science. 1994;266:1697–1702. doi: 10.1126/science.7992053. [DOI] [PubMed] [Google Scholar]
- 10.Gibson G, Hogness D. Science. 1996;271:200–203. doi: 10.1126/science.271.5246.200. [DOI] [PubMed] [Google Scholar]
- 11.Kempin S A, Savidge B, Yanofsky M F. Science. 1995;267:522–525. doi: 10.1126/science.7824951. [DOI] [PubMed] [Google Scholar]
- 12.Bowman J, Alvarez J, Weigel D, Meyerowitz E M, Smyth D R. Development (Cambridge, U.K.) 1993;119:721–743. [Google Scholar]
- 13.Purugganan M D. J Mol Evol. 1997;45:392–396. doi: 10.1007/pl00006244. [DOI] [PubMed] [Google Scholar]
- 14.Ausubel F, editor. Short Protocols in Molecular Biology. New York: Wiley; 1992. [Google Scholar]
- 15.Swofford D. Phylogenetic Analysis Using Parsimony 3.1. Champaign, IL: Illinois Natural History Survey; 1992. [Google Scholar]
- 16.Kumar S, Tamura K, Nei M. Molecular Evolutionary Genetic Analysis Package 1.1. Pennsylvania State University, State College, PA: Institute of Molecular Evolutionary Genetics; 1994. [Google Scholar]
- 17.Hey J, Wakeley J. Genetics. 1997;145:833–846. doi: 10.1093/genetics/145.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tajima F. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fu Y X, Li W H. Genetics. 1993;133:693–709. doi: 10.1093/genetics/133.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simonsen K L, Churchill G A, Aquadro C F. Genetics. 1995;141:413–429. doi: 10.1093/genetics/141.1.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hudson R R, Kaplan N L. Genetics. 1985;111:147–164. doi: 10.1093/genetics/111.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Innan H, Tajima F, Terauchi R, Miyashita N. Genetics. 1996;143:1761–1770. doi: 10.1093/genetics/143.4.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moriyama E N, Powell J R. Mol Biol Evol. 1996;13:261–277. doi: 10.1093/oxfordjournals.molbev.a025563. [DOI] [PubMed] [Google Scholar]
- 24.McDonald J H, Kreitman M. Nature (London) 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- 25.Kamabe A, Innan H, Terauchi R, Miyashita N T. Mol Biol Evol. 1997;14:1303–1315. doi: 10.1093/oxfordjournals.molbev.a025740. [DOI] [PubMed] [Google Scholar]
- 26.Ohta T. Bioessays. 1996;18:673–677. doi: 10.1002/bies.950180811. [DOI] [PubMed] [Google Scholar]
- 27.Ohta T. Annu Rev Ecol Syst. 1992;23:263–286. [Google Scholar]
- 28.Nachman M W, Boyer S N, Aquadro C F. Proc Natl Acad Sci USA. 1994;91:6364–6368. doi: 10.1073/pnas.91.14.6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ballard J W, Kreitman M. Genetics. 1994;138:757–772. doi: 10.1093/genetics/138.3.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Charlesworth B, Nordborg M, Charlesworth D. Genet Res. 1997;70:155–174. doi: 10.1017/s0016672397002954. [DOI] [PubMed] [Google Scholar]
- 31.Rozas J, Rozas R. CABIOS. 1997;13:307–311. [PubMed] [Google Scholar]