Abstract
The X protein (HBX) of the hepatitis B virus (HBV) has been shown to be important for the establishment of HBV infection in vivo. Our previous studies suggested that interaction of HBX with the proteasome complex may underlie the pleiotropic functions of HBX. In this study, we generated a series of woodchuck hepatitis virus (WHV) X mutants, including mutants of the domain interacting with the proteasome, and studied their infectivity in woodchucks. Many of the mutants were defective in transactivation but none of them were completely replication defective in vitro. In vivo, all the wild-type and some X mutant–transfected animals demonstrated evidence of infection with anti-WHc and/or anti-WHs seroconversion. Most of the wild-type– and X mutant–transfected animals had transient viremia. Some animals were later challenged with infectious WHV. Animals inoculated with X mutants, including those with no serologic evidence of infection, were protected from the challenge, suggesting previous infection with resulting protective immunity. Our study demonstrates that the previously described functional domains of HBX are biologically important and the X-defective mutants, possibly as attenuated viruses, are not completely replication defective in vivo.
Introduction
Human hepatitis B virus (HBV) is a member of the Hepadnaviridae family, which includes the hepatitis viruses of the woodchuck, ground squirrel, tree squirrel, Pekin duck, and heron. HBV has a unique fourth open reading frame, termed the hepatitis B virus X (HBX) gene. HBX gene is well conserved among the mammalian hepadnaviruses and codes for a 16.5-kDa protein. The protein can activate the transcription of a variety of viral and cellular genes (1, 2) and induce liver cancer in certain transgenic mouse models (3). Since HBX does not bind to DNA directly, its activity is thought to be mediated by protein-protein interactions. HBX has been shown to enhance transcription through AP-1 and AP-2 (4, 5) and to activate various signal transduction pathways (6, 7). Several recent studies have also identified possible cellular targets of HBX, including members of the CREB/ATF family (8), the TATA-binding protein (9), RNA polymerase subunit RPB5 (10), the ultraviolet-damaged (UV-damaged) DNA-binding protein (11), and the replicative senescence protein p55sen (12). HBX has also been shown to interact with p53 and inhibit its function (13, 14).
Using the Saccharomyces cerevisiae two-hybrid system and a modified yeast two-hybrid dissociator system, we have recently identified two proteasome subunits — PSMA7 (an α proteasome subunit) and PSMC1 (an ATPase-like member of the 19S regulatory factors) — as a putative cellular target of HBX (15, 16). We demonstrated that this interaction may be functionally important in the pleiotropic effect of HBX (17). Because woodchuck hepatitis virus (WHV), like HBV, encodes an X protein (WHVX) with transactivation function and WHVX also interacts with the two proteasome subunits (15), infection of woodchucks would be a valuable model for studying the WHVX function in vivo. In this study, we created a series of site-directed WHVX mutants, including mutants of the domain interacting with the proteasome subunits, and studied the in vivo behaviors of these mutants in woodchucks.
Methods
Plasmid construction.
A 1.2× full-length genome of an infectious WHV strain (pWHV8) was used in this study (18). Various WHVX mutants (Figure 1) were generated by PCR mutagenesis (QuickChange Site-Directed Mutagenesis Kit; Strategene, La Jolla, California, USA) and confirmed by sequencing: WHV8Xlg– with ATG→TTG mutation at the first translation initiation site of WHVX, WHV8Xmd–, and WHV8Xsm– with ATG→GTG mutations at the second and third in-frame translation initiation sites, respectively; WHV8G67S with Gly→Ser at amino acid (aa) 67, WHV8P68A with Pro→Ala at aa 68, WHV8G132V with Gly→Val at aa 132, and WHV8H135D with His→Asp at aa 135. The WHV8XRN– was created by introducing a new in-frame TTG codon to ATG at aa 55 in the Xlg– construct. These X mutations do not cause any amino acid changes in the polymerase gene overlapping the X gene. A replication-defective WHV control, WHV8Pmt, was created by mutating the Asp-Asp→Ala-Ala at the YMDD motif of the polymerase gene. The introduction of these two mutations has been shown previously to inactivate the pol gene (19, 20).
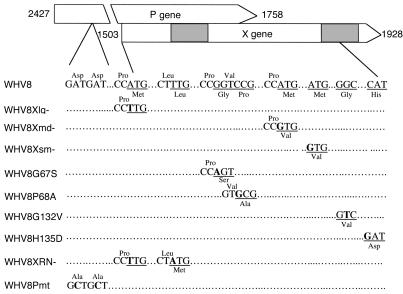
Figure 1.
Schematic diagram of WHVX mutants. The WHVX mutants are displayed in reference to the genomic map of WHV8. The shadow regions represent the two important structural domains of WHVX. The mutations and their approximate positions are shown. The nucleotide sequences are shown in the middle, the amino acids of the mutations are shown at the bottom for the X gene and at the top for the overlapping pol gene (P gene).
DNA transfection and transactivation assay.
Luciferase (Luc) reporter construct RSV-Luc consisting of Rous sarcoma virus promoter driving the luciferase gene was used for WHVX transactivation assays in the HepG2 cells as described previously (15, 16). The human hepatoma HepG2 or HuH7 cells were grown in DMEM (Life Technologies Inc., Rockville, Maryland, USA) containing 10% FBS in a humidified incubator (5% CO2). Transient transfection of HepG2 cells in a 35-mm well was carried out using the DNA transfection kit (5 prime→3 prime Inc., Boulder, Colorado, USA). Luciferase assay was performed with a Monolight Luminometer (Analytical Luminescence Laboratory, San Diego, California, USA). All transfections were controlled for transfection efficiency by cotransfecting a pTK-GH plasmid as described previously (15).
Viral nucleic acid analyses.
HuH7 or HepG2 cells were transiently transfected with WHV8 clones described above. Three or four days after transfection, the cells were harvested for viral RNA and DNA analyses. RNA was prepared by the guanidinium isothiocyanate-acid-phenol method (ULTRASPEC RNA isolation kit; BIOTECX Laboratories, Houston, Texas, USA), analyzed by 1% formaldehyde agarose gel electrophoresis, and hybridized with [32P]-labeled WHV-specific probe. Viral replicative intermediates associated with intracellular core particles were isolated by ultracentrifugation of cell lysate through a 30% sucrose cushion and analyzed by Southern blot hybridization as described (21).
Intrahepatic inoculation of woodchucks with WHV DNAs and challenge of woodchucks with serum-derived WHV7.
All the experimental woodchucks were offspring of WHV-free parents that were born and raised in the animal facility of the College of Veterinary Medicine at Cornell University. For intrahepatic inoculation of woodchucks, various WHV8 constructs were purified by standard protocols (Plasmid Maxi Kit; QIAGEN Inc., Valencia, California, USA). Three animals were transfected with each mutant, and six animals were inoculated with wild-type WHV8 DNA as positive controls. To avoid cross-contamination, inoculations with wild-type and mutant X constructs were performed on different days, and the animals were kept at different containment rooms. Each woodchuck received a dose of 50 μg plasmid DNA in a volume of 0.5 ml of PBS. Starting 2 weeks after transfection, serum samples were collected biweekly for a total of 24 weeks. Liver biopsies were performed every 4 weeks. At week 24 after transfection, selective woodchucks were challenged with 107 infectious dose of serum-derived WHV7. Serum samples were collected biweekly until 12 weeks after challenge.
Serological and nucleic acid assays.
For markers of WHV infection, serum samples collected serially were tested for the presence of WHsAg and Ab’s against WHV core (anti-WHc) or sAg (anti-WHs) using solid-phase radioimmunoassay or ELISA. WHV DNA was assayed routinely by the slot blot hybridization method. For the detection of low viremia of the transfected woodchucks, WHV virions from woodchuck serum were concentrated by ultracentrifuge through a 30% sucrose cushion and treated with micrococcal nuclease to eliminate contaminating free nucleic acids. WHV DNAs were purified by proteinase K digestion and phenol extraction as described previously (21). The virion DNAs were amplified with WHVX-specific primers by PCR with Herculase Enhanced DNA polymerase (Strategene). Several known negative serum samples were included in each assay, and if any of them became positive, the results were discarded and the assay repeated.
Amplification, cloning, and sequencing of WHV DNA from woodchuck samples.
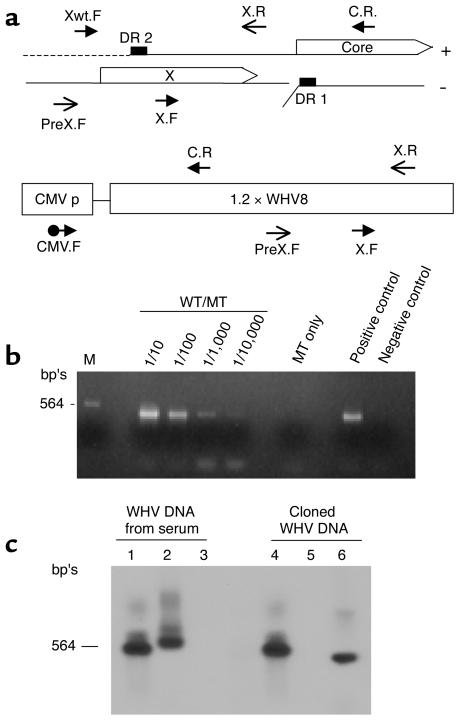
To avoid minor contamination of mutant WHV constructs with wild-type DNAs, the mutant DNAs were produced using disposable glassware at a different laboratory and on different days from that of the wild-type construct. The mutant constructs that were used for inoculation were further analyzed using the amplification refractory mutational detection system, which is based on the absolute inability of an oligonucleotide with mismatched 3′ residue to function as PCR primer under appropriate conditions (22, 23). In brief, an oligonucleotide primer of the wild-type sequence was designed to specifically amplify the wild-type sequence at the particular mutational site. For the Xlg– mutation, the wild-type primer Xwt.F, 5′ AAGCTGACGTCCTTTACA 3′ (nt 1486 to 1503), together with the downstream primer X.R, was designed for the specific detection of wild-type sequence under the PCR condition of 94°C for 30 seconds, 60°C for 30 seconds, and 72°C for 1 minute using the hot-start Pfx DNA polymerase (Life Technology Inc.).
The virion DNAs were purified as described above and amplified with two pairs of WHV primers: the first pair (PreX.F primer, nt 1392 to 1410: 5′ TATTGCCACGGCGGAACTC 3′; X.R primer, nt 1935 to 1914: 5′ TACATGGTTACAGAAGTCGCAT 3′) amplifies the N terminus of WHVX, whereas the second pair (S.F primer, nt 1748 to 1767: 5′ ATTCCACCGTGAACTTTGTC 3′; C.R primer, nt 2346 to 2327: 5′ GTGAGACATGACAAATGAAA 3′) amplifies the C terminus. To distinguish the virion DNA and the cytomegalovirus promoter–driven (CMV-driven) plasmid DNA, a CMV-specific sense primer (CMV.F primer, 5′ AAATGGGCGGTAGGCGTGTA 3′) and the X.R primer were used (see Figure 5). The amplified fragments were cloned into plasmid vector pCR4Blunt-TOPO (Invitrogen Corp., Carlsbad, California, USA) and sequenced.
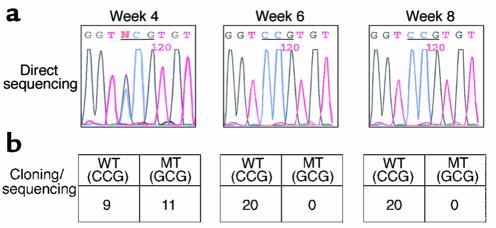
Figure 5.
Sequencing and cloning of PCR product. Serum samples from challenged animals were analyzed by both direct sequencing and cloning followed by sequencing of the PCR products. Direct sequencing (a) and cloning/sequencing (b) of the PCR products from P68A-transfected animal 6017 at weeks 4, 6, and 8 after challenge are shown as an example. The sequences were confirmed by three independent serum extraction/PCR analyses.
Results
Construction of WHV mutants.
HBX has been proposed to contain three in-frame translation initiation sites that may result in multiple polypeptides with different functions (24). WHVX, like HBX, also contains three alternative translation initiation sites. We mutated the three sites separately, resulting in three WHVX mutants, named Xlg–, Xmd–, and Xsm– (Figure 1). Both HBV and WHV X genes contain two important structural and functional domains, with the second domain obligatory for interaction with the proteasome (15, 16). We created two first-domain mutants, G67S and P68A, and two second-domain mutants, G132V and H135D, respectively (Figure 1). We have shown previously that these mutations do not affect the stability of HBX (15). The N terminus of the X gene has been suggested to contain a regulatory domain of about 50 aa (25). To study the function of this domain in vivo, we created a mutant, XRN–, by altering the first ATG to TTG and a TTG codon at aa 55 to ATG, which is in a proper Kozak context for translation initiation codon (26). This mutant was shown to direct the production of a smaller X by in vitro translation (not shown). A replication-defective WHV control, WHV8Pmt, was generated by mutating the Asp-Asp→Ala-Ala at the YMDD motif of the polymerase gene (19, 20).
Transactivation by WHVX mutants.
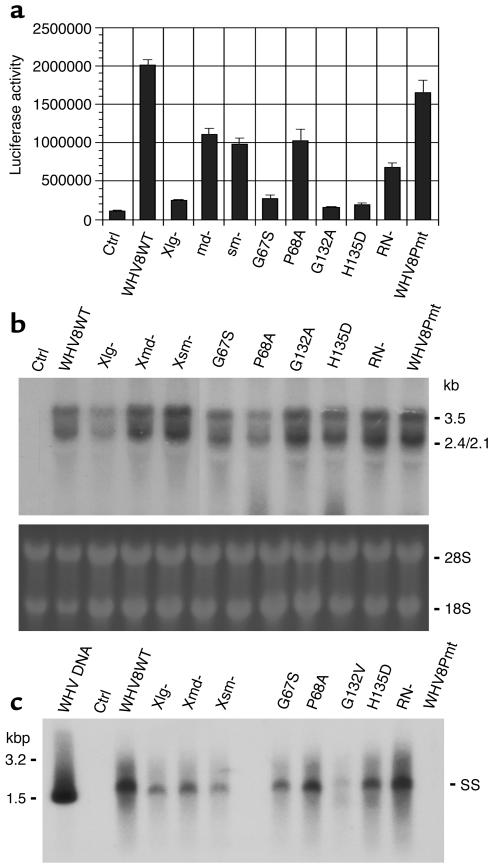
To test the transactivation function of the WHVX mutants, HepG2 cells were cotransfected with the WHV expression constructs and the reporter construct RSV-Luc. The wild-type WHV exerted a 20-fold transactivation of the reporter gene, the WHV8Pmt functioned in a manner similar to the wild-type, and the md–, sm–, P68A, and RN– constructs transactivated at about 30–60% of the wild-type level. All other mutants had little or no transactivation function (Figure 2a).
Figure 2.
Functional characterization of WHVX mutants in vitro. (a) Transactivation of WHVX mutants. The reporter plasmid RSV-Luc and WHVX mutants were cotransfected at a ratio of 1:5 with a total DNA of 0.6 μg per well in a six-well plate. Luciferase activities were assayed 3 days later. Data shown are the mean values of triplicates. (b) Transcription of WHV8X mutants. Three days after transfection, HuH7 cells were harvested, and total RNAs were isolated and subjected to Northern blot analysis. The ribosomal RNAs (bottom) were shown for RNA-loading control (probing for β-actin showed similar results). (c) Replication of WHV8X mutants. Three days after transfection, WHVX-transfected HuH7 cells were harvested, and the core particle–associated WHV DNA were isolated and subjected to Southern blot analysis. Lanes are as indicated. SS, single-stranded WHV DNA. All the results are representative of at least three independent experiments.
Transcription and replication of WHVX mutants.
To evaluate the WHVX mutants at the transcription and replication levels, viral nucleic acids were analyzed in transfected HuH7 cells. Compared with the wild-type WHV8, the viral transcripts of Xlg–, G132V, H135D, and RN– were reduced about 50%. All other mutants appeared to have WHV RNA levels similar to wild-type (Figure 2b). For the analyses of viral replication, viral DNA replicative intermediates associated with intracellular core particles were isolated and analyzed by Southern blot hybridization (Figure 2c). Compared with the wild-type WHV8, the RN– mutant replicated similarly, but all other mutants exhibited variable reductions in the level of replicative intermediates, ranging from about 10% (Xlg–) to 60% (md– and sm–). As expected, the WHV8Pmt mutant showed no replication (Figure 2c).
Infectivity of WHVX mutants in woodchucks.
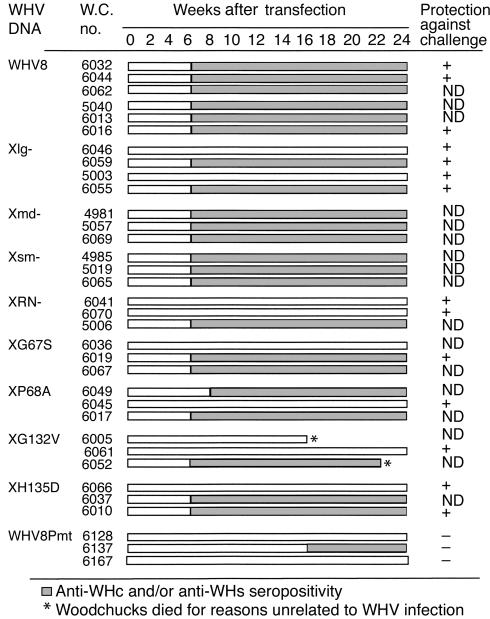
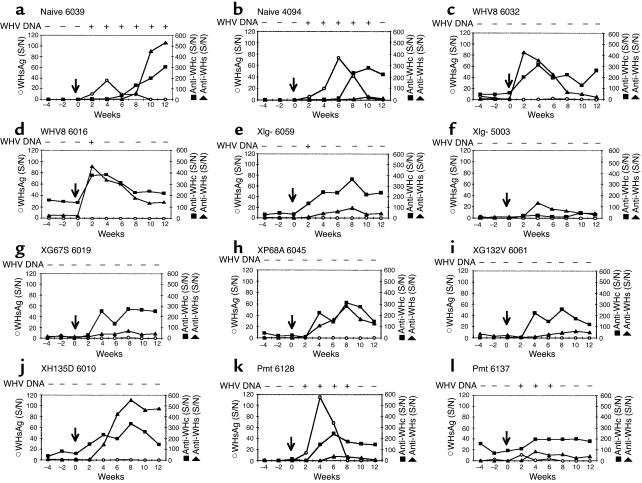
To study the infectivity of WHVX mutants in woodchucks, WHV-free adult woodchucks were transfected with the WHV8X mutants and tested for WHV serological markers and serum WHV DNA posttransfection (Figure 3). All the wild-type–transfected animals demonstrated seroconversion and five of six of the animals had transient low-level viremia. Surprisingly, two of four Xlg–-transfected animals exhibited anti-WHc and/or anti-WHs seroconversion. A fraction of other X mutant–transfected animals also displayed serologic evidence of infection. Most of the X mutant animals also had transient viremia regardless of serologic status. Most surprisingly, one of the three Pmt-transfected woodchucks also developed anti-WHc. However, none of the Pmt-transfected animals had demonstrable viremia (Table 1). Therefore, the presence of anti-WHc in this animal is probably a result of DNA immunization.
Figure 3.
Infectivity of WHVX mutants in woodchucks. WHV-free adult woodchucks were transfected with WHV8X mutants, and serological markers were tested biweekly after transfection. Open bars depict samples with no serological evidence of WHV infection, while filled boxes depict serological evidence of WHV infection with anti-WHc and/or anti-WHs seropositivity. At week 24 after inoculation, selected woodchucks were challenged with 107 WID50 of WHV7 and followed for evidence of protection. The details of the challenge experiments are described in Figure 6, and the results are summarized here. + ,protection; − , no protection; ND, not done.
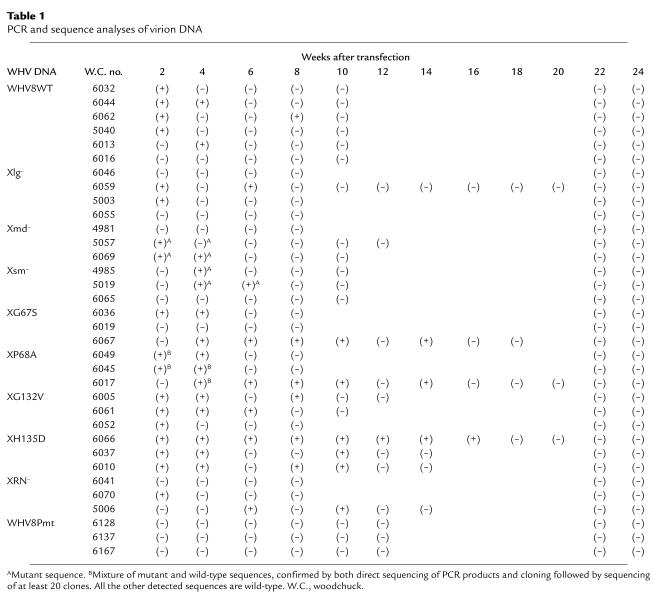
Table 1.
PCR and sequence analyses of virion DNA
Analyses of WHV sequences.
To eliminate the potential contamination of mutant with wild-type WHV construct during the DNA preparation stage, a wild-type–specific primer was designed for the amplification refractory mutation detection system (22, 23). The assay showed that no contamination of wild-type could be detected at a ratio of 1:10,000 (Figure 4b). Because 50 μg of WHV plasmid DNA was injected into the animals, it is possible that any detected WHV DNA in the serum could be a result of this residual DNA. Therefore, we developed a strategy to distinguish between circulating virion DNA and cloned plasmid DNA (Figure 4, a and c). First, we purified the viral particles by ultracentrifugation in a sucrose gradient and then treated the particles with micrococcal nuclease to eliminate all residual contaminating nucleic acids. Control experiments with spiked cloned WHV DNA in the serum showed at least a 6-log depletion of the plasmid DNA using this method (not shown). Second, we developed a selective PCR method to distinguish the two forms of WHV DNAs with three pairs of primers: PreX.F and X.R pair to amplify N terminus of X (NX) in both viral and cloned DNAs, X.F and C.R pair to amplify the C terminus of X (CX) virion DNA only, and CMV.F and C.R pair to amplify cloned DNA (CMV.X) only. The positions of these primers on the viral and cloned DNA were shown in Figure 4a. Figure 4c shows PCR results with these three pairs of primers and different template DNAs followed by Southern blot analyses with WHV DNA probe. When virion DNA was used as the template, both NX and CX PCRs were positive (lanes 1 and 2), while CMVX was negative (lanes 3); when cloned DNA was the template, both NX and CMVX PCRs were positive (lanes 4 and 6), while CX was negative (lanes 5). The sensitivity of detecting circulating virions by this assay is in the range of 103 genomes/ml.
Figure 4.
PCR analyses of WHV DNAs. (a) Standard PCRs were performed with three pairs of primers: Pre-X.F and X.R pair (NX pair) to amplify N terminus of X with expected PCR product of 543 bp’s; X.F and C.R pair (CX pair) to selectively amplify C terminus of X of viral DNA (expected PCR product: 597 bp’s); CMV.F and C.R pair (CMVX pair) to amplify only cloned DNA (expected PCR product: 504 bp’s). The positions of these primers in the viral DNA and cloned DNA are shown. (b) Amplification refractory mutation detection system for wild-type sequence in X mutant inoculum DNA. The position of the wild-type–specific primer Xwt.F is shown in a. Wild-type and Xlg– constructs were mixed at ratios of 1:10 to 1:10,000. One nanogram of each mixture and the mutant inoculum was amplified for 30 cycles as described in Methods. (c) PCR-Southern analyses of viral and cloned WHV DNAs with three pairs of primers: lanes 1 and 4 with NX pair; lanes 2 and 5 with CX pair; lanes 3 and 6 with CMVX pair. The viral DNA was extracted from WHV-containing serum of known titer. About 1 pg of viral or cloned DNA was used for the analyses.
The specific PCR products from woodchuck sera were cloned and sequenced and are summarized in Table 1. Five of six wild-type–transfected animals had detectable wild-type sequences between week 2 to 8. Two of four Xlg-transfected animals had detectable viremia between week 2 to 6, but all the sequences were of the wild-type sequence. Two of the three animals in both Xmd– and Xsm– groups had circulating viral DNA, and all the sequences contained the appropriate mutations. The viral DNA from XP68A-transfected animals had a mixture of mutant and wild-type sequences at weeks 2 and 4, but then became completely wild-type sequences (Figure 5). In all the other X mutant–transfected animals, only wild-type sequences were detected in the serum.
Challenge with infectious WHV.
At 24 weeks after transfection, selective woodchucks were challenged with a 107 infectious dose of serum-derived WHV7. The serum samples were collected and tested for serological evidence of protection against WHV infection. The data are summarized in Figure 3. All the animals with positive serology were protected from the challenge: wild-type animals 6032, 6044, and 6016; Xlg– 6059 and 6055; G67S 6019; and H135D 6010. Surprisingly, even woodchucks with no serologic evidence of infection (Xlg– 6046 and 5003, XRN– 6041 and 6070, P68A 6045, G132V 6061, and H135D 6066) were protected, suggesting establishment of low-level infection with immunologic priming. Two animals (WHV8 6016 and Xlg– 6059) had weakly positive WHV DNA as determined by PCR in serum at postchallenge week 2 only, which is likely a result of carryover from the inoculating dose. Because 107 WID50 of WHV (about 108 genomes) was inoculated, a small amount of WHV might remain 2 weeks later. If one assumes the half-life of free virion in circulation is 1–2 days (27, 28), approximately 103 genomes/ml should remain after 2 weeks (∼10 half-lives), which is at the detection limit of our assay. We also challenged four naive animals (animals 2962, 2956, 4039, and 4094), and three Pmt-inoculated animals (animals 6128, 6137, and 6167). Both naives and Pmt animals were not protected against the challenge and developed transient viremia with positive WHsAg and WHV DNA in serum, despite the presence of anti-WHc Ab’s in Pmt 6137. Detailed serological profiles of some of the challenged animals are summarized in Figure 6.
Figure 6.
Challenge of the transfected woodchucks with WHV7. At 24 weeks after transfection, selective woodchucks were challenged with a 107 infectious dose of serum-derived WHV7. Serological markers were tested biweekly from 4 weeks before challenge (weeks –4 and –2) to 12 weeks after challenge. Time 0 with an arrow represents the time of challenge. The serologic profiles of some of the challenged animals are shown. Signal-to-noise (mean OD of three serum samples of the animal before intrahepatic inoculation) ratio (S/N) for each serologic assay is shown. S/N greater than 3 is considered positive. Serum WHV DNA was assayed as described in Methods, and duplicate assays were performed independently for each serum sample. Only samples positive for both assays were scored as positive. For sample with only one positive assay, a third was performed; if the third was positive, the sample was scored as positive.
Discussion
Because of the essential role of the X gene in the life cycle of mammalian hepadnaviruses in vivo, characterization of the structural and functional properties of X protein in an infectious animal model is crucial. While the avian hepadnavirus may encode an X-equivalent gene (29), the X gene of WHV has been studied in the woodchuck, which probably represents the best small animal model to address the issue. We have demonstrated previously the structural and functional interactions between HBX and the proteasome complex and characterized the domain of HBX that is important for this interaction with a well-defined structural and functional correlation (15–17). In this study, we introduced various single amino acid mutations into the domains of HBX that have been shown to be functionally important for X in vitro, including the domain interacting with the proteasome. In adult woodchucks, these X mutants appeared to behave like attenuated viruses with low-level replication and anti-WHV seroconversion in some animals. Interestingly, these animals were protected from high-dose challenge of infectious WHV, despite the absence of anti-WHV Ab’s in some animals. They all exhibited anamnestic Ab response after the challenge. A WHV mutant with mutations inactivating the pol gene was constructed as a control and was replication defective in woodchucks. Surprisingly, one of three developed anti-WHc Ab’s, which was likely a result of DNA immunization, but none of them were protected from the challenge (Figure 3 and 6). Given the small amount of DNA injected, the lack of protection is consistent with previous studies on DNA immunization in woodchucks (30).
Previous studies of WHVX mutants have shown that they were noninfectious in vivo (31, 32). The study by Chen et al. (31) used a WHV construct that is self-ligated monomeric WHV genome, which is likely a less-efficient transfecting construct in vivo than the CMV-driven terminally redundant WHV plasmid used in our study. The advantage of the latter construct is that it does not require any manipulation such as self-ligation, and it is capable of directing a first-round synthesis of pregenomic RNA independent of the endogenous HBV promoter. This construct has also been used to test the effect of X mutations in woodchucks, and the study also failed to show any infectivity of the mutant (32). However, in the study of Zoulim et al., only one animal was tested for the X mutant, and in our study we tested four animals. Furthermore, the above two studies depended on serology for infectivity determination and did not look for low-level viremia by PCR. As shown in our study, several animals inoculated with WHVX mutants did not develop anti-WHV Ab’s but had demonstrable viremia. Finally the challenge study, performed only in our study, further supported the interpretation that X-deficient mutant can replicate at low level in vivo.
Analyses of WHV sequences during the acute phase of infection revealed the reversion of X mutants to the wild-type sequence except in the animals inoculated with the Xmd– and Xsm– mutants. Although Xmd– and Xsm– mutants had diminished transactivation functions and replicated at a reduced efficiency in cell culture, they seemed to behave similarly to the wild-type in vivo. Therefore, the notion of alternative initiations of X gene may not be biologically relevant (24). However, there may be subtle phenotypic differences that could not be discerned in this study. In animals inoculated with other X mutants, the genotypic reversion to wild-type supports the interpretation that early replication of the X mutants did occur in the woodchucks; otherwise the wild-type revertant would not have a chance to emerge. The temporal sequence of the reversion of X mutants to wild-type occurred rather quickly during the first 2 weeks (Table 1). Most of the X mutant–inoculated animals had predominantly the wild-type sequence in circulation by week 2. Only the animals inoculated with X mutant, XP68A, demonstrated a mixture of mutant and wild-type sequences in the first few weeks, which was then replaced completely by the wild-type sequence in circulation (Figure 5 and Table 1). Although a minor population of mutant species might still exist, we could not detect them based on the sensitivity of our assay (< 1 of 20).
Based on the calculation of the rate of replication in vivo and the mutational frequency of the error-prone reverse transcriptase, a high probability of the emergence of mutants could be predicted in vivo (28). At the peak of hepadnaviral infection, all possible single nucleotide changes could occur in one day. However, the emergence of a viral species is determined by many factors, one of which is the replication fitness of the viral species (28, 33). Because the wild-type replicates more efficiently than the X mutants, it is expected that the wild-type can quickly replace the mutants, especially during the early phase of infection, in which there is little competition for replication space (34). It is not clear why only the XP68A persisted longer than the other X mutants in vivo. This X mutation may confer a less deleterious effect on the in vivo replication of WHV, as reflected in its partially active transactivation function in vitro (Figure 2a). This possible explanation awaits further experimentation.
The X mutants, while replicate with an attenuated phenotype, are capable of priming the immune response to protect the animals from subsequent challenge. All the animals demonstrated an anamnestic response with a rapid rise of anti-WHs and/or anti-WHc titers. Although the responses in general were not as robust as those in animals inoculated with the wild-type, some X mutant animals did exhibit a rather brisk rise to high titers of anti-WHV Ab’s as the wild-type animals (Figure 6). On the other hand, one animal inoculated with the pol mutant with an anti-WHc response exhibited weak, if any, anamnestic response (anti-WHc titer rose from 20 to 40 four weeks after challenge). This evidence lends further credence to the interpretation that X-deficient mutant is not completely replication defective and behaves as an attenuated virus in vivo.
A paper published recently has also examined the role of various WHVX mutants in vivo, in correlation with the interaction of X with another cellular factor UVDDB (35). The mutations were introduced into a region (aa 70–100) that corresponds to the interaction domain between X and UVDDB. Similar to our study, the authors also described a phenotype of attenuated infection with reversion to wild-type associated with some of the mutants. However, it is not clear why viremia and genotypic reversion occurred much later (more than 10 weeks) in those X mutant inoculated animals. No longitudinal analyses were performed on these animals to examine the emergence of wild-type from the mutant species. Furthermore, the lack of a completely X-defective mutant (such as the Xlg–) in their study does not permit a phenotypic comparison of the various X mutants with a X-null mutant. In our study, the Xlg– mutant behaved similarly to all the other X mutants (excluding Xmd– and Xsm–). It is evident from these two studies that several domains of X are functionally important in vivo, but the precise roles of these interacting factors with X and productive viral infection await further experimentation.
Our study suggests that X-deficient virus, as an attenuated virus, could be considered as a vaccine candidate because it is capable of inducing protective immune response in vivo. In addition, the intriguing possibility of HBX as a potential target of antiviral development could greatly expand the armamentarium of anti-HBV therapy, which, to date, only consists of interferon and lamivudine.
Acknowledgments
We wish to thank Robert Purcell for providing the pWHV8 plasmid, and Betty Baldwin and Bud Tennant for excellent technical support and helpful advice during the course of this study. This study is partially supported by NIAID contract N01-AI-05399 via the Hepatitis Animal Network.
Footnotes
Zhensheng Zhang and Nobuyuk Torii contributed equally to this work.
References
- 1.Aufiero B, Schneider RJ. The hepatitis B virus X-gene product trans-activates both RNA polymerase II and III promoters. EMBO J. 1990;9:497–504. doi: 10.1002/j.1460-2075.1990.tb08136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Colgrove R, Simon G, Ganem D. Transcriptional activation of homologous and heterologous genes by the hepatitis B virus X gene product in cells permissive for viral replication. J Virol. 1989;63:4019–4026. doi: 10.1128/jvi.63.9.4019-4026.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim CM, Koike K, Saito I, Miyamura T, Jay G. HBx gene of hepatitis B virus K induces liver cancer in transgenic mice. Nature. 1991;351:317–320. doi: 10.1038/351317a0. [DOI] [PubMed] [Google Scholar]
- 4.Benn J, Su F, Doria M, Schneider RJ. Hepatitis B virus HBx protein induces transcription factor AP-1 by activation of extracellular signal-regulated and c-Jun N-terminal mitogen-activated protein kinases. J Virol. 1996;70:4978–4985. doi: 10.1128/jvi.70.8.4978-4985.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seto E, Mitchell PJ, Yeh TS. Transactivation by the hepatitis B virus X protein depends on AP-2 and other transcription factors. Nature. 1990;344:72–74. doi: 10.1038/344072a0. [DOI] [PubMed] [Google Scholar]
- 6.Cross JC, Wen P, Rutter W. Transactivation by hepatitis B virus X protein is promiscuous and dependent on mitogen-activated cellular serine/threonine kinases. Proc Natl Acad Sci USA. 1993;90:8078–8082. doi: 10.1073/pnas.90.17.8078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doria M, Klein N, Lucito R, Schneider RJ. The hepatitis B virus HBx protein is a dual specificity cytoplasmic activator of Ras and nuclear activator of transcription factors. EMBO J. 1995;14:4747–4757. doi: 10.1002/j.1460-2075.1995.tb00156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maguire HF, Hoeffler JP, Siddiqui A. HBV X protein alters the DNA binding specificity of CREB and ATF-2 by protein-protein interactions. Science. 1991;252:842–844. doi: 10.1126/science.1827531. [DOI] [PubMed] [Google Scholar]
- 9.Qadri I, Maguire HF, Siddiqui A. Hepatitis B virus transactivator protein X interacts with the TATA-binding protein. Proc Natl Acad Sci USA. 1995;92:1003–1007. doi: 10.1073/pnas.92.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheong J, Yi M, Lin Y, Murakami S. Human RPB5, a subunit shared by eukaryotic nuclear RNA polymerases, binds human hepatitis B virus X protein and may play a role in X transactivation. EMBO J. 1995;14:143–150. doi: 10.1002/j.1460-2075.1995.tb06984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sitterlin D, et al. Interaction of the UV-damaged DNA-binding protein with hepatitis B virus X protein is conserved among mammalian hepadnaviruses and restricted to transactivation-proficient X-insertion mutants. J Virol. 1997;71:6194–6199. doi: 10.1128/jvi.71.8.6194-6199.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun BS, Zhu X, Clayton MM, Pan J, Feitelson MA. Identification of a protein isolated from senescent human cells that binds to hepatitis B virus X antigen. Hepatology. 1998;27:228–239. doi: 10.1002/hep.510270135. [DOI] [PubMed] [Google Scholar]
- 13.Truant R, Antunovic J, Greenblatt J, Prives C, Cromlish JA. Direct interaction of the hepatitis B virus HBx protein with p53 leads to inhibition by HBx of p53 response element-directed transactivation. J Virol. 1995;69:1851–1859. doi: 10.1128/jvi.69.3.1851-1859.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang XW, et al. Hepatitis B virus X protein inhibits p53 sequence-specific DNA binding, transcriptional activity, and association with transcription factor ERCC3. Proc Natl Acad Sci USA. 1994;91:2230–2234. doi: 10.1073/pnas.91.6.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang J, Kwong J, Sun EC, Liang TJ. Proteasome complex as a potential cellular target of hepatitis B virus X protein. J Virol. 1996;70:5582–5591. doi: 10.1128/jvi.70.8.5582-5591.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Z, et al. Structural and functional characterization of interaction between hepatitis B virus X protein and the proteasome complex. J Biol Chem. 2000;275:15157–15165. doi: 10.1074/jbc.M910378199. [DOI] [PubMed] [Google Scholar]
- 17.Hu Z, et al. Hepatitis B virus X protein is both a substrate and a potential inhibitor of the proteasome complex. J Virol. 1999;73:7231–7240. doi: 10.1128/jvi.73.9.7231-7240.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Girones MR, et al. Complete nucleotide sequence of a molecular clone of woodchuck hepatitis virus that is infectious in the natural host. Proc Natl Acad Sci USA. 1989;86:1846–1849. doi: 10.1073/pnas.86.6.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Radziwill G, Tucker W, Schaller H. Mutational analysis of the hepatitis B virus P gene product: domain structure and RNase H activity. J Virol. 1990;64:613–620. doi: 10.1128/jvi.64.2.613-620.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang LJ, Hirsch RC, Ganem D, Varmus HE. Effects of insertional and point mutations on the functions of the duck hepatitis B virus polymerase. J Virol. 1990;64:5553–5558. doi: 10.1128/jvi.64.11.5553-5558.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hasegawa K, Huang J, Rogers SA, Blum HE, Liang TJ. Enhanced replication of a hepatitis B virus mutant associated with an epidemic of fulminant hepatitis. J Virol. 1994;68:1651–1659. doi: 10.1128/jvi.68.3.1651-1659.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Newton CR, et al. Analysis of any point mutation in DNA. The amplification refractory mutation system (ARMS) Nucleic Acids Res. 1989;17:2503–2516. doi: 10.1093/nar/17.7.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liang TJ, et al. Hepatitis B virus precore mutation and fulminant hepatitis in the United States. A polymerase chain reaction–based assay for the detection of specific mutation. J Clin Invest. 1994;93:550–555. doi: 10.1172/JCI117006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kwee L, Lucito R, Aufiero B, Schneider RJ. Alternate translation initiation on hepatitis B virus X mRNA produces multiple polypeptides that differentially transactivate class II and III promoters. J Virol. 1992;66:4382–4389. doi: 10.1128/jvi.66.7.4382-4389.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murakami S, Cheong JH, Kaneko S. Human hepatitis virus X gene encodes a regulatory domain that represses transactivation of X protein. J Biol Chem. 1994;269:15118–15123. [PubMed] [Google Scholar]
- 26.Kozak M. Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucleic Acid Res. 1984;12:857–871. doi: 10.1093/nar/12.2.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsiang M, Rooney JF, Toole JJ, Gibbs CS. Biphasic clearance kinetics of hepatitis B virus from patients during adefovir dipivoxil therapy. Hepatology. 1999;29:1863–1869. doi: 10.1002/hep.510290626. [DOI] [PubMed] [Google Scholar]
- 28.Whalley SA, et al. Kinetics of acute hepatitis B virus infection in humans. J Exp Med. 2001;193:847–854. doi: 10.1084/jem.193.7.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang SF, et al. Duck hepatitis B virus expresses a regulatory HBx-like protein from a hidden open reading frame. J Virol. 2001;75:161–170. doi: 10.1128/JVI.75.1.161-170.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu M, et al. Immunization of woodchucks with plasmids expressing woodchuck hepatitis virus (WHV) core antigen and surface antigen suppresses WHV infection. J Virol. 1999;73:281–289. doi: 10.1128/jvi.73.1.281-289.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen H-S, et al. The woodchuck hepatitis X gene is important for establishment of virus infection in woodchucks. J Virol. 1993;67:1218–1226. doi: 10.1128/jvi.67.3.1218-1226.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zoulim F, Saputelli J, Seeger C. Woodchuck hepatitis virus X protein is required for viral infection in vivo. J Virol. 1994;68:2026–2030. doi: 10.1128/jvi.68.3.2026-2030.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Doo E, Liang TJ. Molecular anatomy and pathophysiologic implications of drug resistance in hepatitis B virus infection. Gastroenterology. 2001;120:1000–1008. doi: 10.1053/gast.2001.22454. [DOI] [PubMed] [Google Scholar]
- 34.Zhang YY, Summers J. Low dynamic state of viral competition in a chronic avian hepadnavirus infection. J Virol. 2000;74:5257–5265. doi: 10.1128/jvi.74.11.5257-5265.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sitterlin D, Bergametti F, Tiollais P, Tennant BC, Transy C. Correct binding of viral X protein to UVDDB-p127 cellular protein is critical for efficient infection by hepatitis B viruses. Oncogene. 2000;19:4427–4431. doi: 10.1038/sj.onc.1203770. [DOI] [PubMed] [Google Scholar]