Abstract
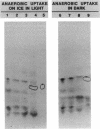
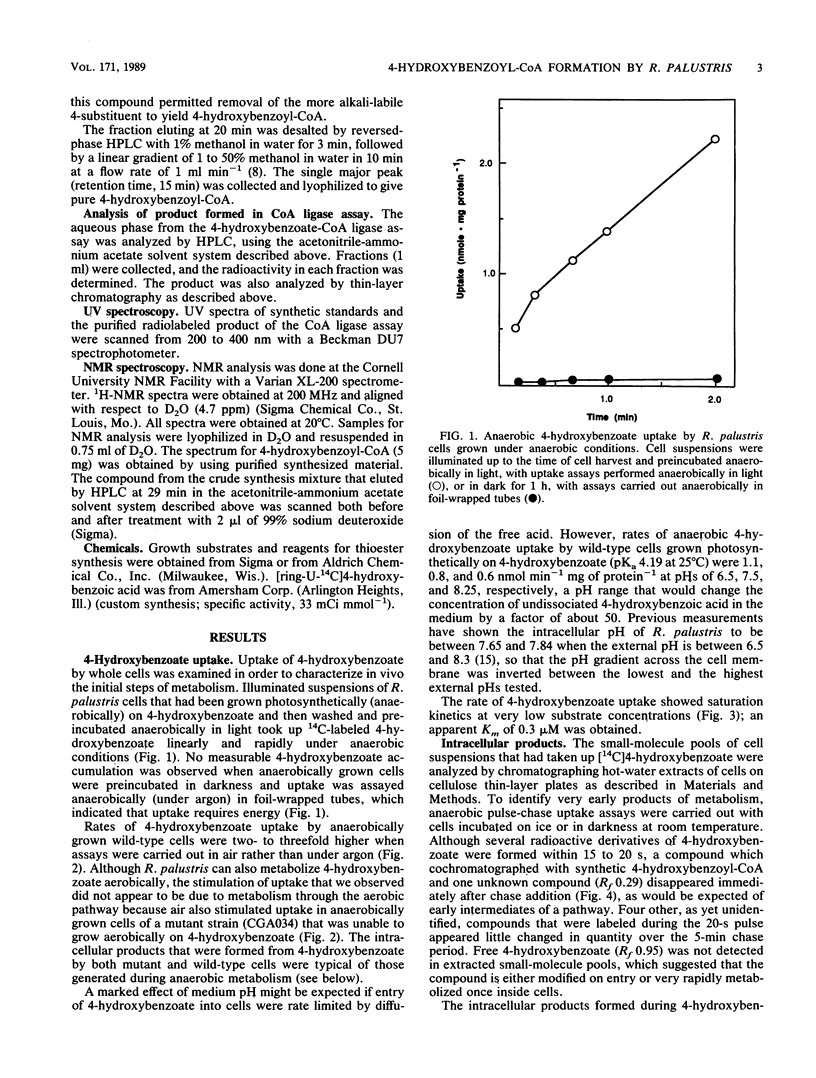
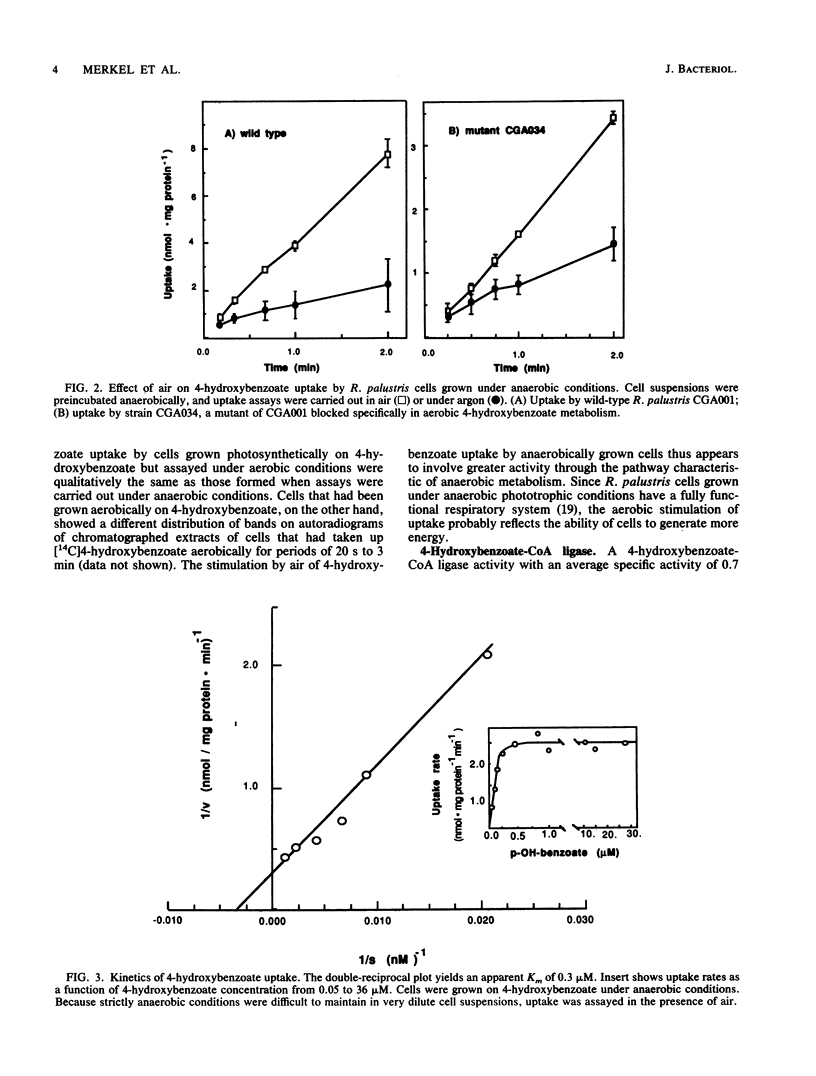
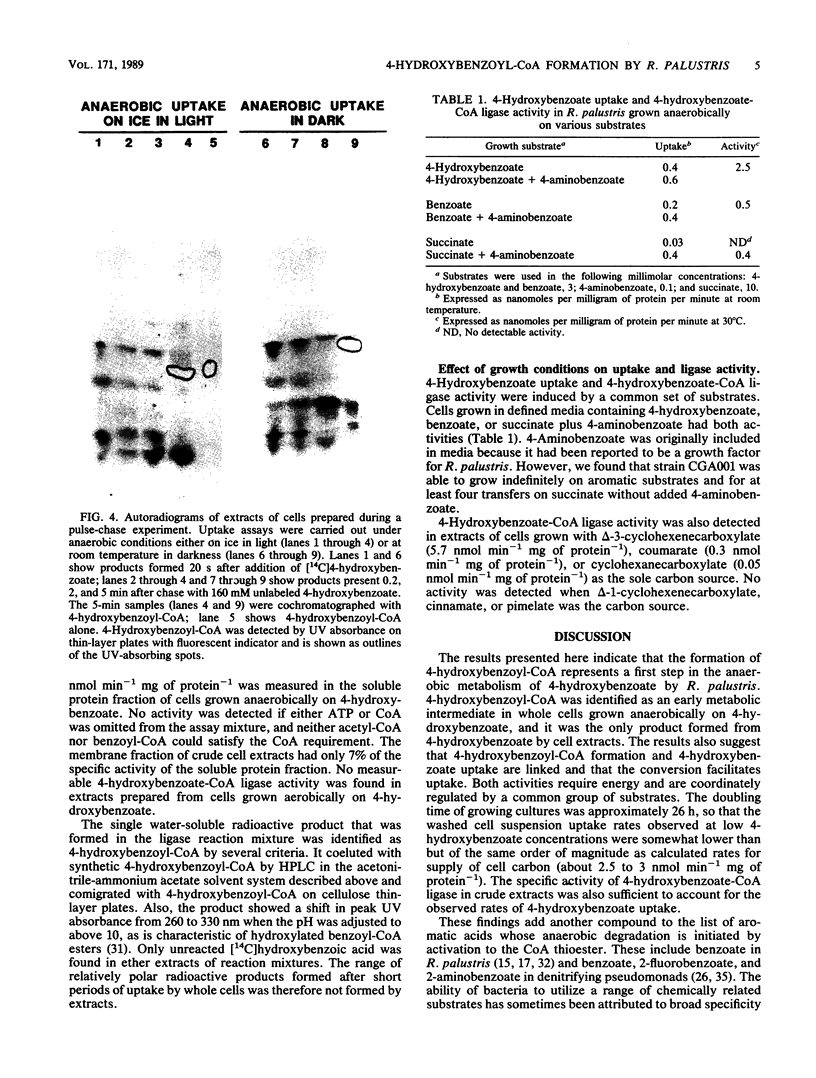
The initial steps of anaerobic 4-hydroxybenzoate degradation were studied in whole cells and cell extracts of the photosynthetic bacterium Rhodopseudomonas palustris. Illuminated suspensions of cells that had been grown anaerobically on 4-hydroxybenzoate and were assayed under anaerobic conditions took up [U-14C]4-hydroxybenzoate at a rate of 0.6 nmol min-1 mg of protein-1. Uptake occurred with high affinity (apparent Km = 0.3 microM), was energy dependent, and was insensitive to external pH in the range of 6.5 to 8.2 Very little free 4-hydroxybenzoate was found associated with cells, but a range of intracellular products was formed after 20-s incubations of whole cells with labeled substrate. When anaerobic pulse-chase experiments were carried out with cells incubated on ice or in darkness, 4-hydroxybenzoyl coenzyme A (4-hydroxybenzoyl-CoA) was formed early and disappeared immediately after addition of excess unlabeled substrate, as would be expected of an early intermediate in 4-hydroxybenzoate metabolism. A 4-hydroxybenzoate-CoA ligase activity with an average specific activity of 0.7 nmol min-1 mg of protein-1 was measured in the soluble protein fraction of cells grown anaerobically on 4-hydroxybenzoate. 4-Hydroxybenzoyl-CoA was the sole product formed from labeled 4-hydroxybenzoate in the ligase reaction mixture. 4-Hydroxybenzoate uptake and ligase activities were present in cells grown anaerobically with benzoate, 4-hydroxybenzoate, and 4-aminobenzoate and were not detected in succinate-grown cells. These results indicate that the high-affinity uptake of 4-hydroxybenzoate by R. palustris is due to rapid conversion of the free acid to its CoA derivative by a CoA ligase and that this is also the initial step of anaerobic 4-hydroxybenzoate degradation.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berry D. F., Francis A. J., Bollag J. M. Microbial metabolism of homocyclic and heterocyclic aromatic compounds under anaerobic conditions. Microbiol Rev. 1987 Mar;51(1):43–59. doi: 10.1128/mr.51.1.43-59.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossert I. D., Young L. Y. Anaerobic oxidation of p-cresol by a denitrifying bacterium. Appl Environ Microbiol. 1986 Nov;52(5):1117–1122. doi: 10.1128/aem.52.5.1117-1122.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Dutton P. L., Evans W. C. The metabolism of aromatic compounds by Rhodopseudomonas palustris. A new, reductive, method of aromatic ring metabolism. Biochem J. 1969 Jul;113(3):525–536. doi: 10.1042/bj1130525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans W. C. Biochemistry of the bacterial catabolism of aromatic compounds in anaerobic environments. Nature. 1977 Nov 3;270(5632):17–22. doi: 10.1038/270017a0. [DOI] [PubMed] [Google Scholar]
- Evans W. C., Fuchs G. Anaerobic degradation of aromatic compounds. Annu Rev Microbiol. 1988;42:289–317. doi: 10.1146/annurev.mi.42.100188.001445. [DOI] [PubMed] [Google Scholar]
- Frazer A. C., Young L. Y. Anaerobic c(1) metabolism of the o-methyl-C-labeled substituent of vanillate. Appl Environ Microbiol. 1986 Jan;51(1):84–87. doi: 10.1128/aem.51.1.84-87.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissler J. F., Harwood C. S., Gibson J. Purification and properties of benzoate-coenzyme A ligase, a Rhodopseudomonas palustris enzyme involved in the anaerobic degradation of benzoate. J Bacteriol. 1988 Apr;170(4):1709–1714. doi: 10.1128/jb.170.4.1709-1714.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grbić-Galić D. Fermentative and oxidative transformation of ferulate by a facultatively anaerobic bacterium isolated from sewage sludge. Appl Environ Microbiol. 1985 Oct;50(4):1052–1057. doi: 10.1128/aem.50.4.1052-1057.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyer M., Hegeman G. Evidence for a reductive pathway for the anaerobic metabolism of benzoate. J Bacteriol. 1969 Sep;99(3):906–907. doi: 10.1128/jb.99.3.906-907.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood C. S., Gibson J. Anaerobic and aerobic metabolism of diverse aromatic compounds by the photosynthetic bacterium Rhodopseudomonas palustris. Appl Environ Microbiol. 1988 Mar;54(3):712–717. doi: 10.1128/aem.54.3.712-717.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood C. S., Gibson J. Uptake of benzoate by Rhodopseudomonas palustris grown anaerobically in light. J Bacteriol. 1986 Feb;165(2):504–509. doi: 10.1128/jb.165.2.504-509.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith C. L., Bridges R. L., Fina L. R., Iverson K. L., Cloran J. A. The anaerobic decomposition of benzoic acid during methane fermentation. IV. Dearomatization of the ring and volatile fatty acids formed on ring rupture. Arch Microbiol. 1978 Aug 1;118(2):173–176. doi: 10.1007/BF00415726. [DOI] [PubMed] [Google Scholar]
- King M. T., Drews G. The respiratory electron transport system of heterotrophically-grown Rhodopseudomonas palustris. Arch Microbiol. 1975 Mar 10;102(3):219–231. doi: 10.1007/BF00428372. [DOI] [PubMed] [Google Scholar]
- Mieyal J. J., Webster L. T., Jr, Siddiqui U. A. Benzoyl and hydroxybenzoyl esters of coenzyme A. Purification and nuclear magnetic resonance characterization; conformation in solution. J Biol Chem. 1974 Apr 25;249(8):2633–2640. [PubMed] [Google Scholar]
- Nunn W. D. A molecular view of fatty acid catabolism in Escherichia coli. Microbiol Rev. 1986 Jun;50(2):179–192. doi: 10.1128/mr.50.2.179-192.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornston L. N., Stanier R. Y. The conversion of catechol and protocatechuate to beta-ketoadipate by Pseudomonas putida. J Biol Chem. 1966 Aug 25;241(16):3776–3786. [PubMed] [Google Scholar]
- Peppercorn M. A., Goldman P. Caffeic acid metabolism by bacteria of the human gastrointestinal tract. J Bacteriol. 1971 Dec;108(3):996–1000. doi: 10.1128/jb.108.3.996-1000.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schennen U., Braun K., Knackmuss H. J. Anaerobic degradation of 2-fluorobenzoate by benzoate-degrading, denitrifying bacteria. J Bacteriol. 1985 Jan;161(1):321–325. doi: 10.1128/jb.161.1.321-325.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor B. F., Campbell W. L., Chinoy I. Anaerobic degradation of the benzene nucleus by a facultatively anaerobic microorganism. J Bacteriol. 1970 May;102(2):430–437. doi: 10.1128/jb.102.2.430-437.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschech A., Fuchs G. Anaerobic degradation of phenol by pure cultures of newly isolated denitrifying pseudomonads. Arch Microbiol. 1987 Sep;148(3):213–217. doi: 10.1007/BF00414814. [DOI] [PubMed] [Google Scholar]
- Webster L. T., Jr, Mieyal J. J., Siddiqui U. A. Benzoyl and hydroxybenzoyl esters of coenzyme A. Ultraviolet characterization and reaction mechanisms. J Biol Chem. 1974 Apr 25;249(8):2641–2645. [PubMed] [Google Scholar]
- Whittle P. J., Lunt D. O., Evans W. C. Anaerobic photometabolism of aromatic compounds by Rhodopseudomonas sp. Biochem Soc Trans. 1976;4(3):490–491. doi: 10.1042/bst0040490. [DOI] [PubMed] [Google Scholar]
- Williams R. J., Evans W. C. The metabolism of benzoate by Moraxella species through anaerobic nitrate respiration. Evidence for a reductive pathway. Biochem J. 1975 Apr;148(1):1–10. doi: 10.1042/bj1480001a. [DOI] [PMC free article] [PubMed] [Google Scholar]