Abstract
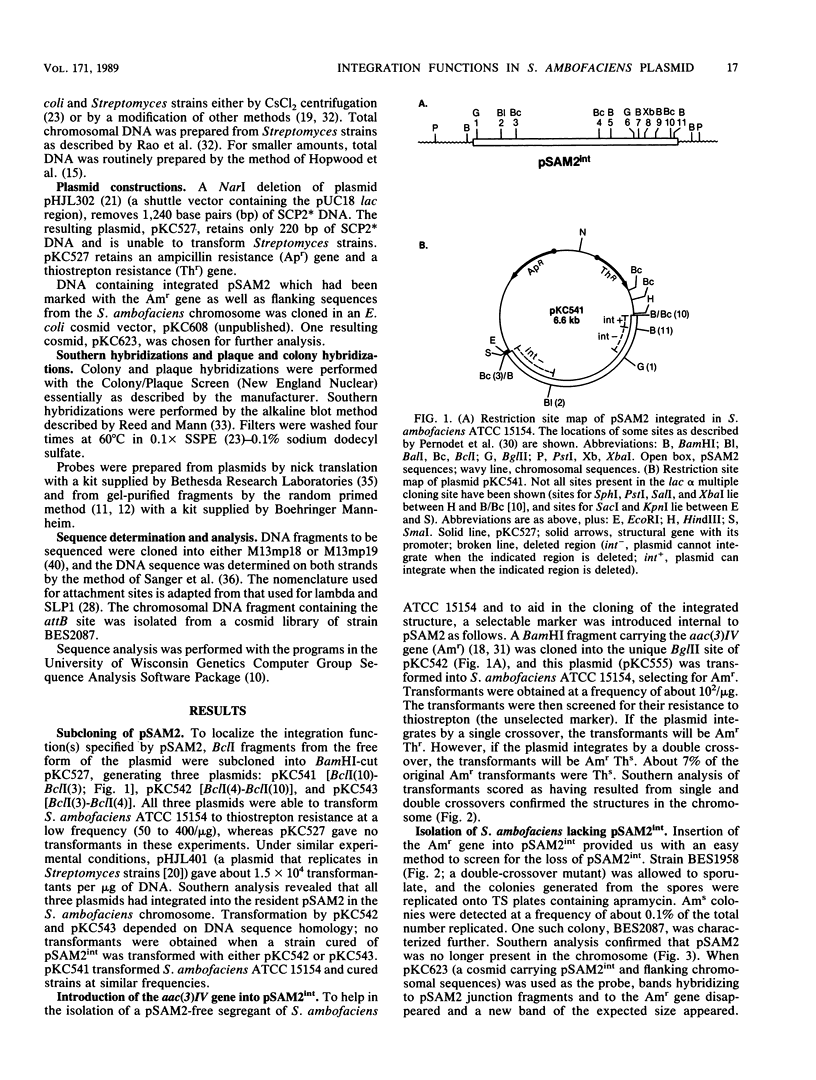
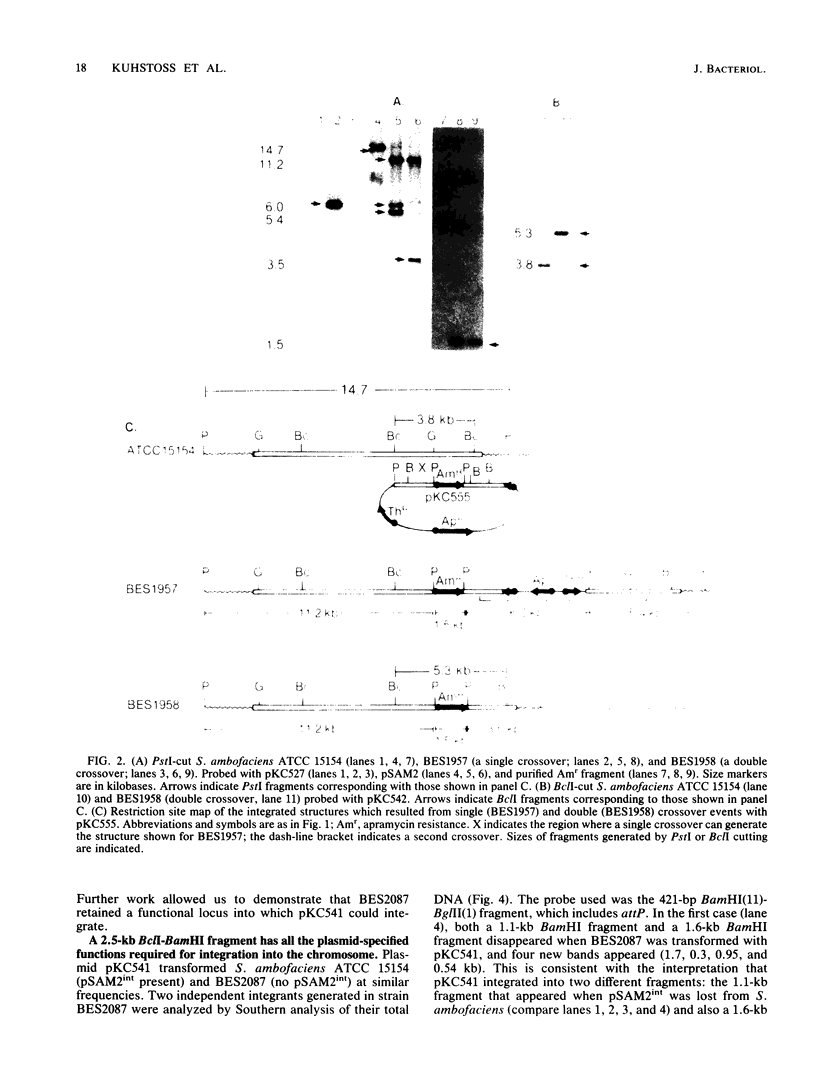
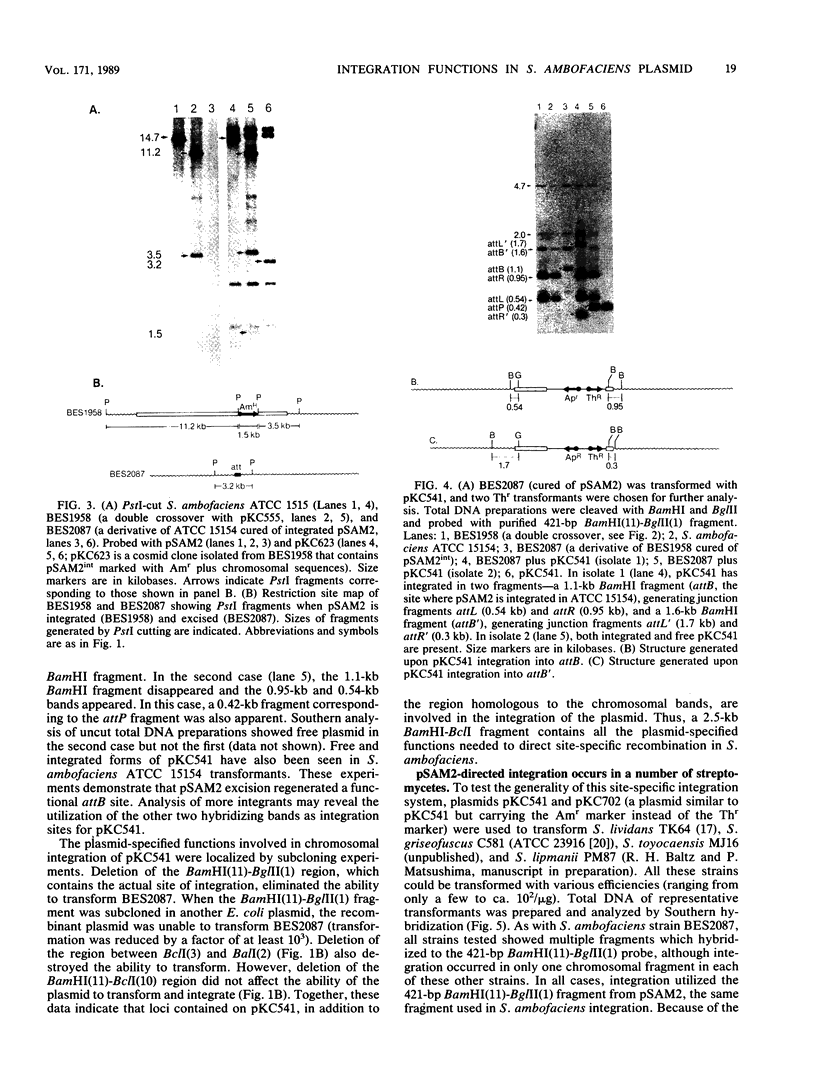
In Streptomyces ambofaciens ATCC 15154, an 11.1-kilobase element, pSAM2, exists as a single integrated copy in the chromosome. In S. ambofaciens 3212 (a derivative of ATCC 15154), pSAM2 exists as a free, circular plasmid as well as an integrated element. BclI fragments from the free form of pSAM2 were cloned into an Escherichia coli plasmid vector. By using gene transplacement methods, the chromosomally integrated form of pSAM2 was marked with a gene coding for apramycin resistance. This enabled us to isolate both a segregant that had lost the integrated pSAM2 element and a cosmid clone containing integrated pSAM2 along with the flanking chromosomal sequences. One of the BclI fragments derived from free pSAM2 was shown to contain all the plasmid-specified information required to direct site-specific recombination in a derivative of S. ambofaciens lacking the resident pSAM2 element as well as in a number of other Streptomyces strains. The attachment sites used by the plasmid and the chromosome in site-specific recombination and the junctions created after integration were cloned and sequenced. Certain structural features in common with other integrating elements in actinomycetes were noted.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andrews B. J., Proteau G. A., Beatty L. G., Sadowski P. D. The FLP recombinase of the 2 micron circle DNA of yeast: interaction with its target sequences. Cell. 1985 Apr;40(4):795–803. doi: 10.1016/0092-8674(85)90339-3. [DOI] [PubMed] [Google Scholar]
- Baltz R. H., Matsushima P. Protoplast fusion in Streptomyces: conditions for efficient genetic recombination and cell regeneration. J Gen Microbiol. 1981 Nov;127(1):137–146. doi: 10.1099/00221287-127-1-137. [DOI] [PubMed] [Google Scholar]
- Bibb M. J., Ward J. M., Kieser T., Cohen S. N., Hopwood D. A. Excision of chromosomal DNA sequences from Streptomyces coelicolor forms a novel family of plasmids detectable in Streptomyces lividans. Mol Gen Genet. 1981;184(2):230–240. doi: 10.1007/BF00272910. [DOI] [PubMed] [Google Scholar]
- Brown D. P., Chiang S. J., Tuan J. S., Katz L. Site-specific integration in Saccharopolyspora erythraea and multisite integration in Streptomyces lividans of actinomycete plasmid pSE101. J Bacteriol. 1988 May;170(5):2287–2295. doi: 10.1128/jb.170.5.2287-2295.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chater K. F., Bruton C. J. Mutational cloning in Streptomyces and the isolation of antibiotic production genes. Gene. 1983 Dec;26(1):67–78. doi: 10.1016/0378-1119(83)90037-9. [DOI] [PubMed] [Google Scholar]
- Cohen A., Bar-Nir D., Goedeke M. E., Parag Y. The integrated and free states of Streptomyces griseus plasmid pSG1. Plasmid. 1985 Jan;13(1):41–50. doi: 10.1016/0147-619x(85)90054-x. [DOI] [PubMed] [Google Scholar]
- Cosloy S. D., Oishi M. Genetic transformation in Escherichia coli K12. Proc Natl Acad Sci U S A. 1973 Jan;70(1):84–87. doi: 10.1073/pnas.70.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox K. L., Baltz R. H. Restriction of bacteriophage plaque formation in Streptomyces spp. J Bacteriol. 1984 Aug;159(2):499–504. doi: 10.1128/jb.159.2.499-504.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. "A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity". Addendum. Anal Biochem. 1984 Feb;137(1):266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Hoess R. H., Abremski K. Mechanism of strand cleavage and exchange in the Cre-lox site-specific recombination system. J Mol Biol. 1985 Feb 5;181(3):351–362. doi: 10.1016/0022-2836(85)90224-4. [DOI] [PubMed] [Google Scholar]
- Hopwood D. A., Hintermann G., Kieser T., Wright H. M. Integrated DNA sequences in three streptomycetes form related autonomous plasmids after transfer to Streptomyces lividans. Plasmid. 1984 Jan;11(1):1–16. doi: 10.1016/0147-619x(84)90002-7. [DOI] [PubMed] [Google Scholar]
- Hopwood D. A., Kieser T., Wright H. M., Bibb M. J. Plasmids, recombination and chromosome mapping in Streptomyces lividans 66. J Gen Microbiol. 1983 Jul;129(7):2257–2269. doi: 10.1099/00221287-129-7-2257. [DOI] [PubMed] [Google Scholar]
- Kieser T. Factors affecting the isolation of CCC DNA from Streptomyces lividans and Escherichia coli. Plasmid. 1984 Jul;12(1):19–36. doi: 10.1016/0147-619x(84)90063-5. [DOI] [PubMed] [Google Scholar]
- Larson J. L., Hershberger C. L. Shuttle vectors for cloning recombinant DNA in Escherichia coli and Streptomyces griseofuscus C581. J Bacteriol. 1984 Jan;157(1):314–317. doi: 10.1128/jb.157.1.314-317.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson J. L., Hershberger C. L. The minimal replicon of a streptomycete plasmid produces an ultrahigh level of plasmid DNA. Plasmid. 1986 May;15(3):199–209. doi: 10.1016/0147-619x(86)90038-7. [DOI] [PubMed] [Google Scholar]
- Madon J., Moretti P., Hütter R. Site-specific integration and excision of pMEA100 in Nocardia mediterranei. Mol Gen Genet. 1987 Sep;209(2):257–264. doi: 10.1007/BF00329651. [DOI] [PubMed] [Google Scholar]
- Matsushima P., Baltz R. H. Efficient plasmid transformation of Streptomyces ambofaciens and Streptomyces fradiae protoplasts. J Bacteriol. 1985 Jul;163(1):180–185. doi: 10.1128/jb.163.1.180-185.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti P., Hintermann G., Hütter R. Isolation and characterization of an extrachromosomal element from Nocardia mediterranei. Plasmid. 1985 Sep;14(2):126–133. doi: 10.1016/0147-619x(85)90072-1. [DOI] [PubMed] [Google Scholar]
- Omer C. A., Cohen S. N. Plasmid formation in Streptomyces: excision and integration of the SLP1 replicon at a specific chromosomal site. Mol Gen Genet. 1984;196(3):429–438. doi: 10.1007/BF00436190. [DOI] [PubMed] [Google Scholar]
- Omer C. A., Cohen S. N. Structural analysis of plasmid and chromosomal loci involved in site-specific excision and integration of the SLP1 element of Streptomyces coelicolor. J Bacteriol. 1986 Jun;166(3):999–1006. doi: 10.1128/jb.166.3.999-1006.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omer C. A., Stein D., Cohen S. N. Site-specific insertion of biologically functional adventitious genes into the Streptomyces lividans chromosome. J Bacteriol. 1988 May;170(5):2174–2184. doi: 10.1128/jb.170.5.2174-2184.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao R. N., Richardson M. A., Kuhstoss S. Cosmid shuttle vectors for cloning and analysis of Streptomyces DNA. Methods Enzymol. 1987;153:166–198. doi: 10.1016/0076-6879(87)53053-1. [DOI] [PubMed] [Google Scholar]
- Reed K. C., Mann D. A. Rapid transfer of DNA from agarose gels to nylon membranes. Nucleic Acids Res. 1985 Oct 25;13(20):7207–7221. doi: 10.1093/nar/13.20.7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson M. A., Mabe J. A., Beerman N. E., Nakatsukasa W. M., Fayerman J. T. Development of cloning vehicles from the Streptomyces plasmid pFJ103. Gene. 1982 Dec;20(3):451–457. doi: 10.1016/0378-1119(82)90214-1. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonet J. M., Boccard F., Pernodet J. L., Gagnat J., Guérineau M. Excision and integration of a self-transmissible replicon of Streptomyces ambofaciens. Gene. 1987;59(1):137–144. doi: 10.1016/0378-1119(87)90274-5. [DOI] [PubMed] [Google Scholar]
- Thompson C. J., Ward J. M., Hopwood D. A. Cloning of antibiotic resistance and nutritional genes in streptomycetes. J Bacteriol. 1982 Aug;151(2):668–677. doi: 10.1128/jb.151.2.668-677.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston F., Chumley F., Fink G. R. Eviction and transplacement of mutant genes in yeast. Methods Enzymol. 1983;101:211–228. doi: 10.1016/0076-6879(83)01016-2. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]