Abstract
Burkitt lymphoma (BL) is an aggressive B‐cell malignancy with endemic, sporadic and immunodeficiency‐associated variants. It has been known for many years that the fundamental transforming event in BL is the translocation of the MYC gene, and the events that bring about this translocation and those that allow cells to survive with the constitutive expression of MYC have been the subject of intense investigation. Epstein–Barr virus (EBV) infection, malaria, immunodeficiency and spontaneous, somatic mutation can all contribute to the origin and maintenance of this cancer and their mechanisms are the subject of this review.
Burkitt lymphoma (BL) can be classified into three forms which differ in geographic distribution and Epstein–Barr virus (EBV) association: endemic (eBL), sporadic (sBL) and HIV‐associated BL (table 1). The hallmark of all BL tumours is the translocation between the MYC gene and one of the immunoglobulin (Ig) heavy or light chain loci. There is a low background incidence of BL worldwide (sBL), which is rarely associated with EBV and accounts for 1–2% of adult lymphoma in Western Europe and America, but eBL is associated with (EBV) in over 95% of cases and is predominant in the equatorial belt of Africa and other parts of the world where malaria is hyperendemic.1,2,3,4 BLs that display an intermediate association with EBV have also been documented in Egypt and Brazil, where up to 87% of tumours are EBV positive5,6,7 and BL occurs in HIV carriers, where tumours can develop prior to the severe immunosuppression coincident with the onset of AIDS. Approximately 30% of such AIDS‐associated tumours are EBV‐positive.8,9
Endemic EBV‐associated BL has an incidence of 5–10/100 000 children and accounts for up to 74% of childhood malignancies in the African equatorial belt.3 In contrast to sBL, which most frequently involves tumours of the abdomen,4 eBL often presents in the jaw or kidneys10,11 but may also occur in the abdomen, ovaries, facial bones and other extranodal sites.1 The cancer has one of the highest cell proliferation rates of any human tumour (doubling time of tumour 24–48 h).12
Histologically, BL cells are monomorphic medium sized cells with round nuclei, a number of nucleoli and abundant cytoplasm. Tumours display a “starry sky” pattern owing to the presence of high numbers of macrophages, which phagocytose apoptotic debris.1 BL tumour cells usually express IgM,13,14,15 B‐cell markers such as CD19, CD20 and CD22 and markers of germinal centre (GC) centroblasts such as CD10, BCL64 and the human germinal centre‐associated lymphoma (HGAL) protein.16
It remains to be firmly established whether eBL originates from a GC‐derived or memory B cell.17,18,19,20,21,22 The cell surface phenotype of BL tumour cells reflects a GC origin but the site of tumour growth is frequently the jaw or ovary, neither of which normally contain GCs. However, the tumour cells have undergone hypermutation,21,23 a feature of the GC reaction during B‐cell activation and differentiation. Moreover, the breakpoint in the Ig gene to which MYC is transferred in eBL occurs at the V(D)J region, suggesting that translocation occurs during V(D)J recombination. The J segments flanking MYC translocated breakpoints typically exhibit deletions and/or additions of base pairs characteristic of normal Ig V(D)J segment rearrangement.24,25 This is a process catalysed by B‐cell specific V(D)J recombinase activating enzymes RAG‐1/2 which are expressed in both pre‐B cells and GC B cells.26,27 In contrast, the chromosomal breakpoint in sBL and HIV‐associated BL occurs most commonly in the class switch region,28 but since both somatic hypermutation and class switching are events that are normally confined to GC B cells and GC centroblast markers are expressed on BL cells, the BL progenitor cells most likely arise from B cells subjected to chromosomal rearrangements in the GC.
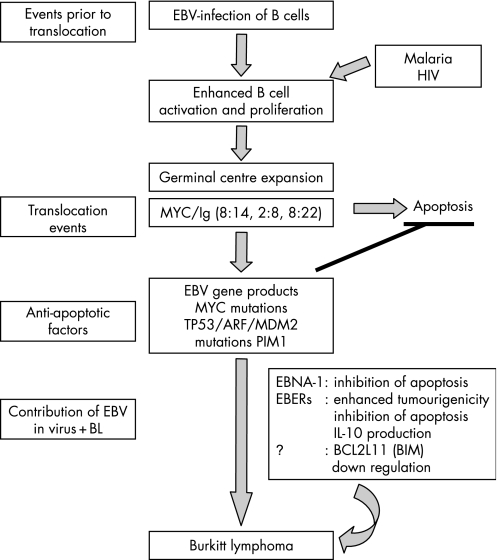
There is some evidence that the cell of origin may be a post‐GC or memory B cell re‐entering the GC18,22 and may differ in EBV‐positive and negative tumours,18 but whichever is the cell of origin, it is clear that GC involvement is critical to the pathogenesis of this disease both in terms of MYC translocation events and the contribution of co‐factors such as EBV, malaria or HIV infection. For example, malaria and HIV infection have both been reported to activate B cells.29,30,31,32 The greater the number of B cells activated and entering the GC reaction the greater the possibility that one cell may subsequently accumulate oncogenic mutations. Interestingly, the C1DR1α motif of the malarial parasite has been shown to drive B‐cell proliferation and protect B cells from apoptosis.32 Furthermore, HIV has been shown to induce the production of cytokines such as interleukin (IL)‐6 and IL‐10 that drive the proliferation of B cells.33,34,35,36 The combination of malaria‐mediated activation and enhanced survival of B cells plus EBV‐driven proliferation of GC B cells may therefore help MYC/Ig translocation‐positive B cells to survive, giving rise to a BL progenitor cell (fig 1).
Figure 1 Pathogenesis of Burkitt lymphoma.
The role of MYC translocations in BL pathogenesis
A defining feature of BL is the reciprocal translocation between the MYC gene and one of the three immunoglobulin genes: the immunoglobulin heavy chain gene (IgH, IGH), and the kappa (IGΚ) or lambda light chain (IGL) genes. In 80% of cases the t(8;14) translocation occurs with the IGH gene. The remaining 20% of cases are split between the translocations with the IGK and IGL (t(2;8) and t(8;22) respectively). Although MYC translocation can also occur in other human cancers such as diffuse large B‐cell lymphoma37 and multiple myeloma,38 it is not thought to be the primary transforming event in these diseases.39,40 Transgenic insertion of MYC into the IgH site resulted in B‐cell and plasma cell neoplasms in mice.41,42,43 Similar translocations have also been observed in the spontaneous cancers murine plasmacytoma and rat immunocytoma, with a predominance of IGH translocations in both cases.44,45
The MYC proto‐oncogene plays a critical role in regulating cell proliferation, differentiation and apoptosis in a cell‐type or context‐dependent manner.46 Its sequence and activities are widely conserved in evolution.47,48,49 The transforming activity of MYC involves its activity as a sequence‐specific transcriptional activator. Its C‐terminal basic helix–loop–helix zipper domain facilitates binding to DNA sequences with the core consensus CACGTG (“E‐boxes”),50 requiring the association of its heterodimeric partner, Max.51 MYC possesses an N‐terminal transactivation domain through which it drives the expression of a large array of target genes,52,53 the mutation of which results in loss of its activity as an oncogene.
The MYC gene comprises three exons. Exon 1 is non‐coding but there are two promoters and regulatory sequences. Exons 2 and 3 contain the protein‐coding sequence beginning on nucleotide 16 of exon 2.54 Most MYC expression occurs from the P2 promoter (80–90% total MYC mRNA),55 but MYC has a complex transcriptional and post‐transcriptional regulation which acts to strictly limit the levels of MYC in the cell. It has a short‐half life and is degraded by ubiquitin‐mediated proteolysis driven first by phosphorylation of serine‐62 followed by threonine‐58, both modifications being activated by the Ras/MAP kinase/Akt pathways.56,57
The breakpoints in the translocated MYC gene occur at different positions in the different forms of BL. In sBL breakpoints are usually within exon 1 or intron 1, whereas the breakpoint in eBL is often at a great distance from the transcriptional start site.58 These differences likely reflect distinct mechanisms of pathogenesis and the differentiation state of target cells in the development of sBL and eBL. However, in both cases, the coding region of the MYC gene is transferred intact. The breakpoint in the immunoglobulin gene to which MYC is transferred also differs in these two forms of BL.
Expression of MYC is normally under tight regulation during the cell cycle but once translocation occurs, expression is constitutive and deregulated, often reaching levels higher than in activated or EBV‐infected B cells. The immunoglobulin enhancers at the adoptive locus appear to be the major deregulating factor on MYC expression, but the locations of breakpoints within MYC have also been correlated with expression levels of the gene product,59 and MYC promoter elements can continue to modulate expression from the new site.60,61,62 Transcription of translocated MYC occurs preferentially from the P1 promoter,63,64 a shift driven by the immunoglobulin enhancers. The normal MYC allele is typically silent in BL,65,66,67 so expression of MYC in these cells is derived solely from the deregulated allele.
Table 1 Characteristic features of the different Burkitt lymphoma subtypes.
| Endemic | Sporadic | HIV‐associated | |
|---|---|---|---|
| Distribution | Equatorial belt of Africa and Papua New Guinea | Worldwide | Worldwide |
| EBV association | 98% | 5−10% | 30−40% |
| Co‐factors | EBV, malaria infection | – | HIV infection |
| Incidence | 5–10/100 000 | 0.01/100 000 | Variable |
| MYC breakpoint | Often >1 kb upstream from 1st coding exon | Exon 1/intron 1 of MYC gene | Exon 1/intron 1 of MYC gene |
| Ig breakpoint | Joining (J) region, switch (S)μ in some cases | Sμ, Sα or J region | Sμ region |
| Progenitor cell | GC, late GC or memory B cell | GC B cell | GC, late GC or memory B cell |
| Frequent site of occurrence | Most frequently jaw. Abdomen, kidneys and ovaries may also be involved | Most frequently abdomen. Kidneys, bone marrow and ovaries may also be involved | Lymph nodes, abdomen, bone marrow, CNS |
Translocation of MYC and the immunoglobulin loci is believed to be aided by the presence of recombination switch sequences in MYC .68,69 Interestingly, higher‐order, spatial organization of B‐cell DNA during interphase puts MYC and the immunoglobulin loci in close proximity, perhaps favouring reciprocal translocations.70 Double, independent MYC translocations have been observed in murine plasmacytoma to both the IGH and IGK or IGL loci, further suggesting a non‐random, reproducible mechanism for MYC transfer.71,72
Genetic translocation is not the only means of MYC deregulation. Mutations which increase the expression, activity and stability of MYC have also been reported.73,74 These mutations are likely to occur after translocation of MYC to the Ig region where somatic hypermutation occurs in germinal centre B cells. Several mutations have been found in the regulatory regions of exon/intron 1 which block negative regulation of MYC expression.75 Mutations in the MYC coding region have also been reported. For example, a commonly occurring mutation in threonine 58 prevents proteolytic degradation of MYC, thereby increasing turnover time of the protein in BL cells.76
Role of other genetic changes in survival of BL tumour cells
A key aspect of MYC function in BL cells is the phenomenon of MYC‐induced apoptosis. While MYC potently drives S phase progression in most somatic cells, cells normally undergo apoptosis when MYC levels exceed a “safe” threshold.77 Over‐expression of MYC in B cells causes induction of p53 or ARF, resulting in apoptosis.78,79 In mouse cells a product of the CDKN2A (INK4a‐ARF) locus, p19ARF80 stabilises p53 by associating with and antagonising MDM2,81,82,83 a key negative regulator of p53.84 High levels of MYC drive apoptosis by inducing p19ARFexpression, resulting in an increase in apoptotic p53.85 In human cells the equivalent (slightly smaller) ARF protein is p14ARF.
Additional changes are therefore selected during the development of BL to counteract this apoptotic effect. The threonine 58 mutation mentioned above also blocks the ability of MYC to induce the expression of the apoptotic BCL‐2 family member BIM. BIM interacts with the anti‐apoptotic protein BCL‐2, inhibiting its function and appears to be an important regulator of apoptosis in these cells.86 Mutations of the T58 site and related amino acids that prevent the phosphorylation of this residue represent an important means by which BL cells retain MYC‐driven proliferation yet evade its apoptotic effects,87 but the exact mechanism by which MYC activates BIM has yet to be described. Recent data shows that EBV‐infected lines express lower levels of BIM than parental lines, suggesting that a latent EBV product blocks apoptosis by down‐regulating the expression of BIM.88,89
Another means of evading MYC‐driven apoptosis is by mutation of TP53, the gene encoding p53. It has long been known that cells lacking p53 and ARF activity are resistant to MYC‐driven apoptosis.90,91 Up to one third of BLs have acquired inactivating TP53 mutations92,93 and most BL cell lines have alterations in some part of the p53/ARF/MDM2 pathway.94 Recently, the anti‐apoptotic kinase, PIM‐1, was reported to be hyperactive in Burkitt lymphoma, causing increased MDM2 levels in these cells, resulting in the destabilisation of p53.95 Restoration of p53 activity in BL cell lines results in a decrease in tumourigenicity.78 Interestingly, BL cells with inactivating TP53 mutations appear to be devoid of MYC mutations.87 These data suggest that once inactivating TP53 mutations have occurred, there is no longer a requirement for further lesions in MYC to block apoptosis.
Mutations that disrupt the nuclear localisation signal of the RB‐related gene, RBL2 (RB2/p130) have also been reported in eBL, correlating with high levels of proliferation.96,97 It was suggested that alterations in p130 may drive proliferation prior to translocation of the MYC gene.
Role of EBV in BL cell survival
The presence of EBV in GC cells that undergo a MYC translocation is also likely to aid cell survival. EBV is a ubiquitous gamma herpesvirus that establishes a seemingly harmless latent infection in B cells in over 95% of the human population, but is also involved in several types of cancer. The identification of clonal EBV genomes in all cells of tumours98 indicates that the progenitor tumour cell was infected with EBV and supports the notion that the virus plays a role at an early stage of tumourigenesis. Moreover, antibodies to the EBV viral capsid antigen (VCA) are raised months or years prior to the development of disease and can correlate with disease burden.99
EBV can display three patterns of latent gene expression: latency I (latency programme), II (default programme) and III (growth programme). Latency III is characterised by expression of all the latent genes (EBNAs, LMPs and EBERS) and occurs on primary infection of B cells, where EBV clearly drives cell proliferation. In contrast, persistent infection in vivo is characterised by expression of EBNA‐1 and LMP‐2 plus the EBER RNAs.100 eBL cells usually express only the EBNA‐1 protein plus the EBERs (latency I), giving rise to debate as to how EBV may directly contribute to tumour growth. One report also detected LMP2A RNA.101
The restricted EBV latent gene expression profile102 and reduced expression of MHC class I, transporter associated with antigen processing (TAP) molecules and the proteasome subunit LMP7 in tumour cells103,104,105 help tumour cells to evade immune surveillance, but EBV gene products also seem to directly aid cell survival in the BL cells. Thus spontaneous loss of the EBV genome during passage of EBV‐positive BL lines in vitro increases their sensitivity to apoptosis. Three per cent of EBV replicons are lost per cell per generation if they do not provide a survival advantage, yet expression of both EBNA‐1 and EBERs is maintained in BL cells.106
In fact, roles for both EBNA‐1 and the EBERs in the prevention of apoptosis and survival of BL cells have been reported. Early studies on transgenic mice expressing EBNA‐1 in B cells suggested a predisposition to develop B cell tumours,107 and experiments performed using a dominant negative EBNA‐1 expressed from retroviral vectors demonstrated that inhibition of EBNA‐1 reduced the survival of EBV‐positive but not EBV‐negative tumour cells in a dose‐dependent manner. Cells in which EBNA‐1 was inhibited displayed a four‐fold increase in the level of apoptosis prior to loss of the EBV genome or changes in the level of the EBERs.108
In EBV‐negative Akata BL cell the EBERs enhanced tumourigenicity and resistance to apoptosis,109,110,111 increasing the growth of tumour cells in soft agar and significantly enhancing the tumourigenicity of EBV‐negative BL cells in SCID mice.110,111 EBERs (or EBER1 alone) have also been reported to bind to and inhibit the dsRNA‐activated protein kinase, PKR112,113,114,115,116 and consequently inhibit IFN‐α induced apoptosis.114 PKR regulates cellular stress and apoptotic pathways, but its reported role as a tumour suppressor led to suggestions that inhibition of its function by EBERs may play a role in tumourigenesis. The direct role of EBER‐mediated PKR inhibition in mediating IFNα‐induced apoptosis resistance has since been challenged,111 casting doubt over a potential role for PKR in BL development under these conditions. Interestingly, EBERs have also been reported to be responsible for increased production of the B‐cell growth factor, IL‐10, in EBV‐positive BL lines compared to EBV‐negative BL lines.117 IL‐10 was shown to be present at higher levels in the tumour microenvironment of EBV‐positive BL compared to EBV negative BL.117,118 The recently discovered microRNAs in EBV do not appear to be expressed at high levels in BL cells,119 but seem likely to be important in some other EBV‐associated diseases such as nasopharyngeal carcinoma.
While EBNA‐1 and the EBERs are generally thought to be the only EBV genes expressed in eBL, recent studies have found that a minor proportion of eBL tumours has a novel form of latency in which the EBNA‐3A, 3B, 3C and LP latent genes are expressed in the absence of EBNA‐2 and LMP‐1 or 2.103 These tumours contain deletion mutants of EBV lacking the EBNA2 gene and this has led to the idea that EBNA2 is incompatible with the de‐regulated MYC expression in BL cells, suggesting a selective pressure for loss of EBNA‐2 expression (either latency I or deletion of EBNA2). Further investigation has demonstrated that eBL tumours may be comprised of tumour cells expressing variable patterns of EBV gene expression, each of which confer a different level of resistance to apoptosis.120 In addition to the EBNA‐2‐deleted virus, EBNA‐2+ LMP‐1− clones and the previously described EBNA‐1 only clones were identified. This finding supports early immunohistochemical studies in which the latent genes LMP‐1 and EBNA‐2 were identified in a proportion of eBL tumour cells.121 Thus, EBNA‐1, 3A, 3B, 3C and LP positive EBNA‐2, LMP negative BL cells were the most resistant to apoptosis, while EBNA‐2+, LMP1‐negative BL cells displayed reduced but “intermediate” resistance. Latency I BL cells displayed the least resistance to apoptosis but afforded some level of protection compared to EBV‐negative BL cells. Reports of eBL cases in Malawi in which EBNA‐1, LMP1, LMP‐2A, BZLF‐1, EBERs and the BARTs were identified122 suggest that EBV gene expression may be broader than previously thought, but these occasional exceptions can be seen as typical tumour heterogeneity and should not detract from our understanding of the usual EBNA1 and EBER only pattern of EBV gene expression in BL.
Malaria as a BL cofactor
The role of malarial infection in the pathogenesis of eBL is clear for the geographical co‐incidence of the two diseases. It is generally thought that the association between malaria and BL arises from a combination of immunosuppression and B‐cell activation. For example, cytotoxic T‐cell mediated control over the outgrowth EBV‐infected B cells is impaired during acute malaria infection,123,124,125 and it has been found that peripheral EBV loads may be five times higher during acute malaria compared to levels observed during convalescence or in healthy individuals.126 EBV loads are generally higher in areas of holoendemic malaria compared to areas where malaria is sporadic,127 and show increased persistence in children with a history of severe rather than mild malaria,128 possibly owing to higher viral reactivation.129 eBL also develops at a later age in individuals who have migrated from malaria‐free high altitude areas to lower, malaria‐endemic areas.130
In support of these findings, it has recently been found that the malarial parasite Plasmodium falciparum can directly activate B cells via a cysteine‐rich interdomain region 1α (C1DR1α) on the P falciparum erythrocyte membrane protein 1 (PfEMP1), which binds to surface Ig. The activation of B cells by C1DR1α and subsequent protection from apoptosis has been postulated to play a role in enhancing survival of GC B cells bearing oncogenic mutations.32 In addition to the activation of B cells, it is possible that proliferation of B cells is enhanced by IL‐10. Serum levels of this cytokine are raised in children suffering from acute P falciparum malaria compared to healthy controls.131
Take‐home messages
The key proliferative change to all Burkitt lymphoma cells is the chromosome translocation of MYC to one of the immunoglobulin loci.
There are many factors that can contribute to stabilising the hyperproliferative, yet apoptotic phenotype that results from MYC overexpression
There are many roles that Epstein–Barr virus can play in both the formation and maintenance of Burkitt lymphoma.
Protective immunity is only acquired following several years of exposure to the malarial parasite P falciparum,132 and the intervening immunosuppression in malaria endemic areas may alter the regulation of EBV‐positive B cells.8,133 Immunosuppression is probably part of the mechanism of HIV‐associated BL, but this can develop prior to the severe loss of immunity characteristic of AIDS, suggesting that severe immunosuppression is not a prerequisite for BL development. Additionally, EBV‐associated tumours in post‐transplant patients, in whom immunosuppression is severe, tend to display a latency III type EBV gene expression profile rather than the restricted pattern frequently seen in eBL patients. In addition to the influence of malaria in stimulating B‐cell expansion, the possibility that mosquito‐borne arboviruses are another risk factor for eBL has recently been raised.3
Conclusion
While BL is undoubtedly a disease of MYC translocation, there are many other pathological factors which occur around this key event. These factors conspire to increase the probability of translocation or to stabilise the hyperproliferative, yet apoptotic phenotype that results from its overexpression. The patterns with which they occur clearly differ between the immunologically and geographically distinct forms of BL and are frequently indicative of the events leading to their respective pathogenesis. There are many roles that EBV can play in both the formation and maintenance of this disease and current research is actively exploring these mechanisms.
Footnotes
Competing interests: None declared.
References
- 1.Ferry J A. Burkitt's lymphoma: clinicopathologic features and differential diagnosis. Oncologist 200611375–383. [DOI] [PubMed] [Google Scholar]
- 2.Pattle S B, Farrell P J. The role of Epstein–Barr virus in cancer. Expert Opin Biol Ther 200661193–1205. [DOI] [PubMed] [Google Scholar]
- 3.van den Bosch C A. Is endemic Burkitt's lymphoma an alliance between three infections and a tumour promoter? Lancet Oncol 20045738–746. [DOI] [PubMed] [Google Scholar]
- 4.Blum K A, Lozanski G, Byrd J C. Adult Burkitt leukemia and lymphoma. Blood 20041043009–3020. [DOI] [PubMed] [Google Scholar]
- 5.Anwar N, Kingma D W, Bloch A R.et al The investigation of Epstein–Barr viral sequences in 41 cases of Burkitt's lymphoma from Egypt: epidemiologic correlations. Cancer 1995761245–1252. [DOI] [PubMed] [Google Scholar]
- 6.Araujo I, Foss H D, Hummel M.et al Frequent expansion of Epstein–Barr virus (EBV) infected cells in germinal centres of tonsils from an area with a high incidence of EBV‐associated lymphoma. J Pathol 1999187326–330. [DOI] [PubMed] [Google Scholar]
- 7.Klumb C E, Hassan R, De Oliveira D E.et al Geographic variation in Epstein–Barr virus‐associated Burkitt's lymphoma in children from Brazil. Int J Cancer 200410866–70. [DOI] [PubMed] [Google Scholar]
- 8.Rochford R, Cannon M J, Moormann A M. Endemic Burkitt's lymphoma: a polymicrobial disease? Nat Rev Microbiol 20053182–187. [DOI] [PubMed] [Google Scholar]
- 9.Powles T, Matthews G, Bower M. AIDS related systemic non‐Hodgkin's lymphoma. Sex Transm Infect 200076335–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sariban E, Donahue A, Magrath I T. Jaw involvement in American Burkitt's Lymphoma. Cancer 1984531777–1782. [DOI] [PubMed] [Google Scholar]
- 11.Mwanda O W. Clinical characteristics of Burkitt's lymphoma seen in Kenyan patients. East Afr Med J 2004S78–S89. [DOI] [PubMed]
- 12.Iversen U, Iversen O H, Bluming A Z.et al Cell kinetics of African cases of Burkitt lymphoma. A preliminary report. Eur J Cancer 19728305–308. [DOI] [PubMed] [Google Scholar]
- 13.Rowe M, Rooney C M, Rickinson A B.et al Distinctions between endemic and sporadic forms of Epstein–Barr virus‐positive Burkitt's lymphoma. Int J Cancer 198535435–441. [DOI] [PubMed] [Google Scholar]
- 14.Rowe M, Rooney C M, Edwards C F.et al Epstein–Barr virus status and tumour cell phenotype in sporadic Burkitt's lymphoma. Int J Cancer 198637367–373. [DOI] [PubMed] [Google Scholar]
- 15.Wiels J, Lenoir G M, Fellous M.et al A monoclonal antibody with anti‐Burkitt lymphoma specificity. I. Analysis of human haematopoietic and lymphoid cell lines. Int J Cancer 198229653–658. [DOI] [PubMed] [Google Scholar]
- 16.Natkunam Y, Lossos I S, Taidi B.et al Expression of the human germinal center‐associated lymphoma (HGAL) protein, a new marker of germinal center B‐cell derivation. Blood 20051053979–3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bellan C, Lazzi S, De Falco G.et al Burkitt's lymphoma: new insights into molecular pathogenesis. J Clin Pathol 200356188–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bellan C, Lazzi S, Hummel M.et al Immunoglobulin gene analysis reveals 2 distinct cells of origin for EBV‐positive and EBV‐negative Burkitt lymphomas. Blood 20051061031–1036. [DOI] [PubMed] [Google Scholar]
- 19.Chapman C J, Mockridge C I, Rowe M.et al Analysis of VH genes used by neoplastic B cells in endemic Burkitt's lymphoma shows somatic hypermutation and intraclonal heterogeneity. Blood 1995852176–2181. [PubMed] [Google Scholar]
- 20.Chapman C J, Zhou J X, Gregory C.et al VH and VL gene analysis in sporadic Burkitt's lymphoma shows somatic hypermutation, intraclonal heterogeneity, and a role for antigen selection. Blood 1996883562–3568. [PubMed] [Google Scholar]
- 21.Chapman C J, Wright D, Stevenson F K. Insight into Burkitt's lymphoma from immunoglobulin variable region gene analysis. Leuk Lymphoma 199830257–267. [DOI] [PubMed] [Google Scholar]
- 22.Isobe K, Tamaru J, Nakamura S.et al VH gene analysis in sporadic Burkitt's lymphoma: somatic mutation and intraclonal diversity with special reference to the tumor cells involving germinal center. Leuk Lymphoma 200243159–164. [DOI] [PubMed] [Google Scholar]
- 23.Tamaru J, Hummel M, Marafioti T.et al Burkitt's lymphomas express VH genes with a moderate number of antigen‐selected somatic mutations. Am J Pathol 19951471398–1407. [PMC free article] [PubMed] [Google Scholar]
- 24.Haluska F G, Finver S, Tsujimoto Y.et al The t(8; 14) chromosomal translocation occurring in B‐cell malignancies results from mistakes in V‐D‐J joining. Nature 1986324158–161. [DOI] [PubMed] [Google Scholar]
- 25.Haluska F G, Tsujimoto Y, Croce C M. The t(8;14) chromosome translocation of the Burkitt lymphoma cell line Daudi occurred during immunoglobulin gene rearrangement and involved the heavy chain diversity region. Proc Natl Acad Sci USA 1987846835–6839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hikida M, Mori M, Takai T.et al Reexpression of RAG‐1 and RAG‐2 genes in activated mature mouse B cells. Science 19962742092–2094. [DOI] [PubMed] [Google Scholar]
- 27.Han S, Zheng B, Schatz D G.et al Neoteny in lymphocytes: Rag1 and Rag2 expression in germinal center B cells. Science 19962742094–2097. [DOI] [PubMed] [Google Scholar]
- 28.Magrath I T. African Burkitt's lymphoma. History, biology, clinical features, and treatment. Am J Pediatr Hematol Oncol 199113222–246. [PubMed] [Google Scholar]
- 29.Lane H C, Masur H, Edgar L C.et al Abnormalities of B‐cell activation and immunoregulation in patients with the acquired immunodeficiency syndrome. N Engl J Med 1983309453–458. [DOI] [PubMed] [Google Scholar]
- 30.Greenwood B M, Vick R M. Evidence for a malaria mitogen in human malaria. Nature 1975257592–594. [DOI] [PubMed] [Google Scholar]
- 31.Gabrielsen A A, Jr, Jensen J B. Mitogenic activity of extracts from continuous cultures of Plasmodium falciparum. Am J Trop Med Hyg 198231441–448. [DOI] [PubMed] [Google Scholar]
- 32.Donati D, Mok B, Chene A.et al Increased B cell survival and preferential activation of the memory compartment by a malaria polyclonal B cell activator. J Immunol 20061773035–3044. [DOI] [PubMed] [Google Scholar]
- 33.Nakajima K, Martinez‐Maza O, Hirano T.et al Induction of IL‐6 (B cell stimulatory factor‐2/IFN‐beta 2) production by HIV. J Immunol 1989142531–536. [PubMed] [Google Scholar]
- 34.Masood R, Zhang Y, Bond M W.et al Interleukin‐10 is an autocrine growth factor for acquired immunodeficiency syndrome‐related B‐cell lymphoma. Blood 1995853423–3430. [PubMed] [Google Scholar]
- 35.Masood R, Lunardi‐Iskandar Y, Moudgil T.et al IL‐10 inhibits HIV‐1 replication and is induced by tat. Biochem Biophys Res Commun 1994202374–383. [DOI] [PubMed] [Google Scholar]
- 36.Breen E C, Rezai A R, Nakajima K.et al Infection with HIV is associated with elevated IL‐6 levels and production. J Immunol 1990144480–484. [PubMed] [Google Scholar]
- 37.Ueda C, Nishikori M, Kitawaki T.et al Coexistent rearrangements of c‐MYC, BCL2, and BCL6 genes in a diffuse large B‐cell lymphoma. Int J Hematol 20047952–54. [DOI] [PubMed] [Google Scholar]
- 38.Avet‐Loiseau H, Gerson F, Magrangeas F.et al Rearrangements of the c‐myc oncogene are present in 15% of primary human multiple myeloma tumors. Blood 2001983082–3086. [DOI] [PubMed] [Google Scholar]
- 39.Kanda‐Akano Y, Nomura K, Fujita Y.et al Molecular‐cytogenetic characterization of non‐Hodgkin's lymphoma with double and cryptic translocations of the immunoglobulin heavy chain gene. Leuk Lymphoma 2004451559–1567. [DOI] [PubMed] [Google Scholar]
- 40.Shou Y, Martelli M L, Gabrea A.et al Diverse karyotypic abnormalities of the c‐myc locus associated with c‐myc dysregulation and tumor progression in multiple myeloma. Proc Natl Acad Sci USA 200097228–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adams J M, Harris A W, Pinkert C A.et al The c‐myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature 1985318533–538. [DOI] [PubMed] [Google Scholar]
- 42.Nussenzweig M C, Schmidt E V, Shaw A C.et al A human immunoglobulin gene reduces the incidence of lymphomas in c‐Myc‐bearing transgenic mice. Nature 1988336446–450. [DOI] [PubMed] [Google Scholar]
- 43.Park S S, Kim J S, Tessarollo L.et al Insertion of c‐Myc into Igh induces B‐cell and plasma‐cell neoplasms in mice. Cancer Res 2005651306–1315. [DOI] [PubMed] [Google Scholar]
- 44.Shen‐Ong G L, Keath E J, Piccoli S P.et al Novel myc oncogene RNA from abortive immunoglobulin‐gene recombination in mouse plasmacytomas. Cell 198231443–452. [DOI] [PubMed] [Google Scholar]
- 45.Pear W S, Wahlstrom G, Nelson S F.et al 6;7 chromosomal translocation in spontaneously arising rat immunocytomas: evidence for c‐myc breakpoint clustering and correlation between isotypic expression and the c‐myc target. Mol Cell Biol 19888441–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eisenman R N. Deconstructing myc. Genes Dev 2001152023–2030. [DOI] [PubMed] [Google Scholar]
- 47.Persson H, Hennighausen L, Taub R.et al Antibodies to human c‐myc oncogene product: evidence of an evolutionarily conserved protein induced during cell proliferation. Science 1984225687–693. [DOI] [PubMed] [Google Scholar]
- 48.Sarid J, Halazonetis T D, Murphy W.et al Evolutionarily conserved regions of the human c‐myc protein can be uncoupled from transforming activity. Proc Natl Acad Sci USA 198784170–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schreiber‐Agus N, Alland L, Muhle R.et al A biochemical and biological analysis of Myc superfamily interactions. Curr Top Microbiol Immunol 1997224159–168. [DOI] [PubMed] [Google Scholar]
- 50.Prendergast G C, Ziff E B. A new bind for Myc. Trends Genet 1992891–96. [DOI] [PubMed] [Google Scholar]
- 51.Blackwood E M, Eisenman R N. Max: a helix‐loop‐helix zipper protein that forms a sequence‐specific DNA‐binding complex with Myc. Science 19912511211–1217. [DOI] [PubMed] [Google Scholar]
- 52.Coller H A, Grandori C, Tamayo P.et al Expression analysis with oligonucleotide microarrays reveals that MYC regulates genes involved in growth, cell cycle, signaling, and adhesion. Proc Natl Acad Sci USA 2000973260–3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Z, Van Calcar S, Qu C.et al A global transcriptional regulatory role for c‐Myc in Burkitt's lymphoma cells. Proc Natl Acad Sci USA 20031008164–8169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Spencer C A, Groudine M. Control of c‐myc regulation in normal and neoplastic cells. Adv Cancer Res 1991561–48. [DOI] [PubMed] [Google Scholar]
- 55.Taub R, Moulding C, Battey J.et al Activation and somatic mutation of the translocated c‐myc gene in Burkitt lymphoma cells. Cell 198436339–348. [DOI] [PubMed] [Google Scholar]
- 56.Sears R, Leone G, DeGregori J.et al Ras enhances Myc protein stability. Mol Cell 19993169–179. [DOI] [PubMed] [Google Scholar]
- 57.Sears R, Nuckolls F, Haura E.et al Multiple Ras‐dependent phosphorylation pathways regulate Myc protein stability. Genes Dev 2000142501–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shiramizu B, Barriga F, Neequaye J.et al Patterns of chromosomal breakpoint locations in Burkitt's lymphoma: relevance to geography and Epstein–Barr virus association. Blood 1991771516–1526. [PubMed] [Google Scholar]
- 59.Wilda M, Busch K, Klose I.et al Level of MYC overexpression in pediatric Burkitt's lymphoma is strongly dependent on genomic breakpoint location within the MYC locus. Genes Chromosomes Cancer 200441178–182. [DOI] [PubMed] [Google Scholar]
- 60.Ji L, Arcinas M, Boxer L M. NF‐kappa B sites function as positive regulators of expression of the translocated c‐myc allele in Burkitt's lymphoma. Mol Cell Biol 1994147967–7974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ji L, Arcinas M, Boxer L M. The transcription factor, Nm23H2, binds to and activates the translocated c‐myc allele in Burkitt's lymphoma. J Biol Chem 199527013392–13398. [DOI] [PubMed] [Google Scholar]
- 62.Hortnagel K, Mautner J, Strobl L J.et al The role of immunoglobulin kappa elements in c‐myc activation. Oncogene 1995101393–1401. [PubMed] [Google Scholar]
- 63.Strobl L J, Eick D. Hold back of RNA polymerase II at the transcription start site mediates down‐regulation of c‐myc in vivo. Embo J 1992113307–3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Strobl L J, Kohlhuber F, Mautner J.et al Absence of a paused transcription complex from the c‐myc P2 promoter of the translocation chromosome in Burkitt's lymphoma cells: implication for the c‐myc P1/P2 promoter shift. Oncogene 199381437–1447. [PubMed] [Google Scholar]
- 65.ar‐Rushdi A, Nishikura K, Erikson J.et al Differential expression of the translocated and the untranslocated c‐myc oncogene in Burkitt lymphoma. Science 1983222390–393. [DOI] [PubMed] [Google Scholar]
- 66.Hayday A C, Gillies S D, Saito H.et al Activation of a translocated human c‐myc gene by an enhancer in the immunoglobulin heavy‐chain locus. Nature 1984307334–340. [DOI] [PubMed] [Google Scholar]
- 67.Nishikura K, ar‐Rushdi A, Erikson J.et al Differential expression of the normal and of the translocated human c‐myc oncogenes in B cells. Proc Natl Acad Sci USA 1983804822–4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dunnick W, Shell B E, Dery C. DNA sequences near the site of reciprocal recombination between a c‐myc oncogene and an immunoglobulin switch region. Proc Natl Acad Sci USA 1983807269–7273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gerondakis S, Cory S, Adams J M. Translocation of the myc cellular oncogene to the immunoglobulin heavy chain locus in murine plasmacytomas is an imprecise reciprocal exchange. Cell 198436973–982. [DOI] [PubMed] [Google Scholar]
- 70.Roix J J, McQueen P G, Munson P J.et al Spatial proximity of translocation‐prone gene loci in human lymphomas. Nat Genet 200334287–291. [DOI] [PubMed] [Google Scholar]
- 71.Wiener F, Silva S, Sugiyama H.et al The “missing” mouse plasmacytoma (MPC) associated translocation T(15;16) occurs repeatedly in new MPC induction systems. Genes Chromosomes Cancer 1990236–43. [DOI] [PubMed] [Google Scholar]
- 72.Silva S, Kovalchuk A L, Kim J S.et al BCL2 accelerates inflammation‐induced BALB/c plasmacytomas and promotes novel tumors with coexisting T(12;15) and T(6;15) translocations. Cancer Res 2003638656–8663. [PubMed] [Google Scholar]
- 73.Cesarman E, Dalla‐Favera R, Bentley D.et al Mutations in the first exon are associated with altered transcription of c‐myc in Burkitt lymphoma. Science 19872381272–1275. [DOI] [PubMed] [Google Scholar]
- 74.Yu B W, Ichinose I, Bonham M A.et al Somatic mutations in c‐myc intron I cluster in discrete domains that define protein binding sequences. J Biol Chem 199326819586–19592. [PubMed] [Google Scholar]
- 75.Zajac‐Kaye M, Gelmann E P, Levens D. A point mutation in the c‐myc locus of a Burkitt lymphoma abolishes binding of a nuclear protein. Science 19882401776–1780. [DOI] [PubMed] [Google Scholar]
- 76.Gregory M A, Hann S R. c‐Myc proteolysis by the ubiquitin‐proteasome pathway: stabilization of c‐Myc in Burkitt's lymphoma cells. Mol Cell Biol 2000202423–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Packham G, Cleveland J L. c‐Myc and apoptosis. Biochim Biophys Acta 1995124211–28. [DOI] [PubMed] [Google Scholar]
- 78.Cherney B W, Bhatia K G, Sgadari C.et al Role of the p53 tumor suppressor gene in the tumorigenicity of Burkitt's lymphoma cells. Cancer Res 1997572508–2515. [PubMed] [Google Scholar]
- 79.Eischen C M, Weber J D, Roussel M F.et al Disruption of the ARF‐Mdm2‐p53 tumor suppressor pathway in Myc‐induced lymphomagenesis. Genes Dev 1999132658–2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Quelle D E, Zindy F, Ashmun R A.et al Alternative reading frames of the INK4a tumor suppressor gene encode two unrelated proteins capable of inducing cell cycle arrest. Cell 199583993–1000. [DOI] [PubMed] [Google Scholar]
- 81.Kamijo T, Weber J D, Zambetti G.et al Functional and physical interactions of the ARF tumor suppressor with p53 and Mdm2. Proc Natl Acad Sci USA 1998958292–8297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pomerantz J, Schreiber‐Agus N, Liegeois N J.et al The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2's inhibition of p53. Cell 199892713–723. [DOI] [PubMed] [Google Scholar]
- 83.Stott F J, Bates S, James M C.et al The alternative product from the human CDKN2A locus, p14(ARF), participates in a regulatory feedback loop with p53 and MDM2. Embo J 1998175001–5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Barak Y, Juven T, Haffner R.et al mdm2 expression is induced by wild type p53 activity. Embo J 199312461–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zindy F, Eischen C M, Randle D H.et al Myc signaling via the ARF tumor suppressor regulates p53‐dependent apoptosis and immortalization. Genes Dev 1998122424–2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.O'Connor L, Strasser A, O'Reilly L A.et al Bim: a novel member of the Bcl‐2 family that promotes apoptosis. Embo J 199817384–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hemann M T, Bric A, Teruya‐Feldstein J.et al Evasion of the p53 tumour surveillance network by tumour‐derived MYC mutants. Nature 2005436807–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Clybouw C, McHichi B, Mouhamad S.et al EBV infection of human B lymphocytes leads to down‐regulation of Bim expression: relationship to resistance to apoptosis. J Immunol 20051752968–2973. [DOI] [PubMed] [Google Scholar]
- 89.Leao M, Anderton E, Wade M.et al Epstein–Barr virus‐induced resistance to drugs that activate the mitotic spindle assembly checkpoint in Burkitt's lymphoma cells. J Virol 200781248–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Harvey D M, Levine A J. p53 alteration is a common event in the spontaneous immortalization of primary BALB/c murine embryo fibroblasts. Genes Dev 199152375–2385. [DOI] [PubMed] [Google Scholar]
- 91.Zindy F, Quelle D E, Roussel M F.et al Expression of the p16INK4a tumor suppressor versus other INK4 family members during mouse development and aging. Oncogene 199715203–211. [DOI] [PubMed] [Google Scholar]
- 92.Farrell P J, Allan G J, Shanahan F.et al p53 is frequently mutated in Burkitt's lymphoma cell lines. Embo J 1991102879–2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gaidano G, Ballerini P, Gong J Z.et al p53 mutations in human lymphoid malignancies: association with Burkitt lymphoma and chronic lymphocytic leukemia. Proc Natl Acad Sci USA 1991885413–5417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lindstrom M S, Klangby U, Wiman K G. p14ARF homozygous deletion or MDM2 overexpression in Burkitt lymphoma lines carrying wild type p53. Oncogene 2001202171–2177. [DOI] [PubMed] [Google Scholar]
- 95.Ionov Y, Le X, Tunquist B J.et al Pim‐1 protein kinase is nuclear in Burkitt's lymphoma: nuclear localization is necessary for its biologic effects. Anticancer Res 200323167–178. [PubMed] [Google Scholar]
- 96.Cinti C, Leoncini L, Nyongo A.et al Genetic alterations of the retinoblastoma‐related gene RB2/p130 identify different pathogenetic mechanisms in and among Burkitt's lymphoma subtypes. Am J Pathol 2000156751–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cinti C, Claudio P P, Howard C M.et al Genetic alterations disrupting the nuclear localization of the retinoblastoma‐related gene RB2/p130 in human tumor cell lines and primary tumors. Cancer Res 200060383–389. [PubMed] [Google Scholar]
- 98.Neri A, Barriga F, Inghirami G.et al Epstein–Barr virus infection precedes clonal expansion in Burkitt's and acquired immunodeficiency syndrome‐associated lymphoma. Blood 1991771092–1095. [PubMed] [Google Scholar]
- 99.Pagano J S, Blaser M, Buendia M A.et al Infectious agents and cancer: criteria for a causal relation. Semin Cancer Biol 200414453–471. [DOI] [PubMed] [Google Scholar]
- 100.Thorley‐Lawson D A. Epstein–Barr virus: exploiting the immune system. Nat Rev Immunol 2001175–82. [DOI] [PubMed] [Google Scholar]
- 101.Tao Q, Robertson K D, Manns A.et al Epstein–Barr virus (EBV) in endemic Burkitt's lymphoma: molecular analysis of primary tumor tissue. Blood 1998911373–1381. [PubMed] [Google Scholar]
- 102.Rowe M, Rowe D T, Gregory C D.et al Differences in B cell growth phenotype reflect novel patterns of Epstein–Barr virus latent gene expression in Burkitt's lymphoma cells. Embo J 198762743–2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kelly G, Bell A, Rickinson A. Epstein–Barr virus‐associated Burkitt lymphomagenesis selects for downregulation of the nuclear antigen EBNA2. Nat Med 200281098–1104. [DOI] [PubMed] [Google Scholar]
- 104.Rowe M, Khanna R, Jacob C A.et al Restoration of endogenous antigen processing in Burkitt's lymphoma cells by Epstein–Barr virus latent membrane protein‐1: coordinate up‐regulation of peptide transporters and HLA‐class I antigen expression. Eur J Immunol 1995251374–1384. [DOI] [PubMed] [Google Scholar]
- 105.Khanna R, Burrows S R, Argaet V.et al Endoplasmic reticulum signal sequence facilitated transport of peptide epitopes restores immunogenicity of an antigen processing defective tumour cell line. Int Immunol 19946639–645. [DOI] [PubMed] [Google Scholar]
- 106.Hammerschmidt W, Sugden B. Epstein–Barr virus sustains Burkitt's lymphomas and Hodgkin's disease. Trends Mol Med 200410331–336. [DOI] [PubMed] [Google Scholar]
- 107.Wilson J B, Bell J L, Levine A J. Expression of Epstein–Barr virus nuclear antigen‐1 induces B cell neoplasia in transgenic mice. Embo J 1996153117–3126. [PMC free article] [PubMed] [Google Scholar]
- 108.Kennedy G, Komano J, Sugden B. Epstein–Barr virus provides a survival factor to Burkitt's lymphomas. Proc Natl Acad Sci USA 200310014269–14274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Komano J, Sugiura M, Takada K. Epstein–Barr virus contributes to the malignant phenotype and to apoptosis resistance in Burkitt's lymphoma cell line Akata. J Virol 1998729150–9156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Komano J, Maruo S, Kurozumi K.et al Oncogenic role of Epstein–Barr virus‐encoded RNAs in Burkitt's lymphoma cell line Akata. J Virol 1999739827–9831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ruf I K, Rhyne P W, Yang C.et al Epstein–Barr virus small RNAs potentiate tumorigenicity of Burkitt lymphoma cells independently of an effect on apoptosis. J Virol 20007410223–10228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Nanbo A, Yoshiyama H, Takada K. Epstein–Barr virus‐encoded poly(A)‐ RNA confers resistance to apoptosis mediated through Fas by blocking the PKR pathway in human epithelial intestine 407 cells. J Virol 20057912280–12285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sharp T V, Schwemmle M, Jeffrey I.et al Comparative analysis of the regulation of the interferon‐inducible protein kinase PKR by Epstein–Barr virus RNAs EBER‐1 and EBER‐2 and adenovirus VAI RNA. Nucleic Acids Res 1993214483–4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Nanbo A, Inoue K, Adachi‐Takasawa K.et al Epstein–Barr virus RNA confers resistance to interferon‐alpha‐induced apoptosis in Burkitt's lymphoma. Embo J 200221954–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Elia A, Laing K G, Schofield A.et al Regulation of the double‐stranded RNA‐dependent protein kinase PKR by RNAs encoded by a repeated sequence in the Epstein–Barr virus genome. Nucleic Acids Res 1996244471–4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.McKenna S A, Kim I, Liu C W.et al Uncoupling of RNA binding and PKR kinase activation by viral inhibitor RNAs. J Mol Biol 20063581270–1285. [DOI] [PubMed] [Google Scholar]
- 117.Kitagawa N, Goto M, Kurozumi K.et al Epstein–Barr virus‐encoded poly(A)(‐) RNA supports Burkitt's lymphoma growth through interleukin‐10 induction. Embo J 2000196742–6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ogden C A, Pound J D, Batth B K.et al Enhanced apoptotic cell clearance capacity and B cell survival factor production by IL‐10‐activated macrophages: implications for Burkitt's lymphoma. J Immunol 20051743015–3023. [DOI] [PubMed] [Google Scholar]
- 119.Cai X, Schafer A, Lu S.et al Epstein–Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLoS Pathog 20062e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kelly G L, Milner A E, Baldwin G S.et al Three restricted forms of Epstein–Barr virus latency counteracting apoptosis in c‐myc‐expressing Burkitt lymphoma cells. Proc Natl Acad Sci USA 200610314935–14940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Niedobitek G, Agathanggelou A, Rowe M.et al Heterogeneous expression of Epstein–Barr virus latent proteins in endemic Burkitt's lymphoma. Blood 199586659–665. [PubMed] [Google Scholar]
- 122.Xue S A, Labrecque L G, Lu Q L.et al Promiscuous expression of Epstein–Barr virus genes in Burkitt's lymphoma from the central African country Malawi. Int J Cancer 200299635–643. [DOI] [PubMed] [Google Scholar]
- 123.Moss D J, Burrows S R, Castelino D J.et al A comparison of Epstein–Barr virus‐specific T‐cell immunity in malaria‐endemic and ‐nonendemic regions of Papua New Guinea. Int J Cancer 198331727–732. [DOI] [PubMed] [Google Scholar]
- 124.Whittle H C, Brown J, Marsh K.et al The effects of Plasmodium falciparum malaria on immune control of B lymphocytes in Gambian children. Clin Exp Immunol 199080213–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Whittle H C, Brown J, Marsh K.et al T‐cell control of Epstein–Barr virus‐infected B cells is lost during P falciparum malaria. Nature 1984312449–450. [DOI] [PubMed] [Google Scholar]
- 126.Lam K M, Syed N, Whittle H.et al Circulating Epstein–Barr virus‐carrying B cells in acute malaria. Lancet 1991337876–878. [DOI] [PubMed] [Google Scholar]
- 127.Moormann A M, Chelimo K, Sumba O P.et al Exposure to holoendemic malaria results in elevated Epstein–Barr virus loads in children. J Infect Dis 20051911233–1238. [DOI] [PubMed] [Google Scholar]
- 128.Yone C L, Kube D, Kremsner P G.et al Persistent Epstein–Barr viral reactivation in young African children with a history of severe Plasmodium falciparum malaria. Trans R Soc Trop Med Hyg 2006100669–676. [DOI] [PubMed] [Google Scholar]
- 129.Rasti N, Falk K I, Donati D.et al Circulating Epstein–Barr virus in children living in malaria‐endemic areas. Scand J Immunol 200561461–465. [DOI] [PubMed] [Google Scholar]
- 130.Burkitt D P. The discovery of Burkitt's lymphoma. Cancer 1983511777–1786. [DOI] [PubMed] [Google Scholar]
- 131.Lyke K E, Burges R, Cissoko Y.et al Serum levels of the proinflammatory cytokines interleukin‐1 beta (IL‐1beta), IL‐6, IL‐8, IL‐10, tumor necrosis factor alpha, and IL‐12(p70) in Malian children with severe Plasmodium falciparum malaria and matched uncomplicated malaria or healthy controls. Infect Immun 2004725630–5637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Artavanis‐Tsakonas K, Tongren J E, Riley E M. The war between the malaria parasite and the immune system: immunity, immunoregulation and immunopathology. Clin Exp Immunol 2003133145–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Crawford D H. Biology and disease associations of Epstein–Barr virus. Philos Trans R Soc Lond B Biol Sci 2001356461–473. [DOI] [PMC free article] [PubMed] [Google Scholar]