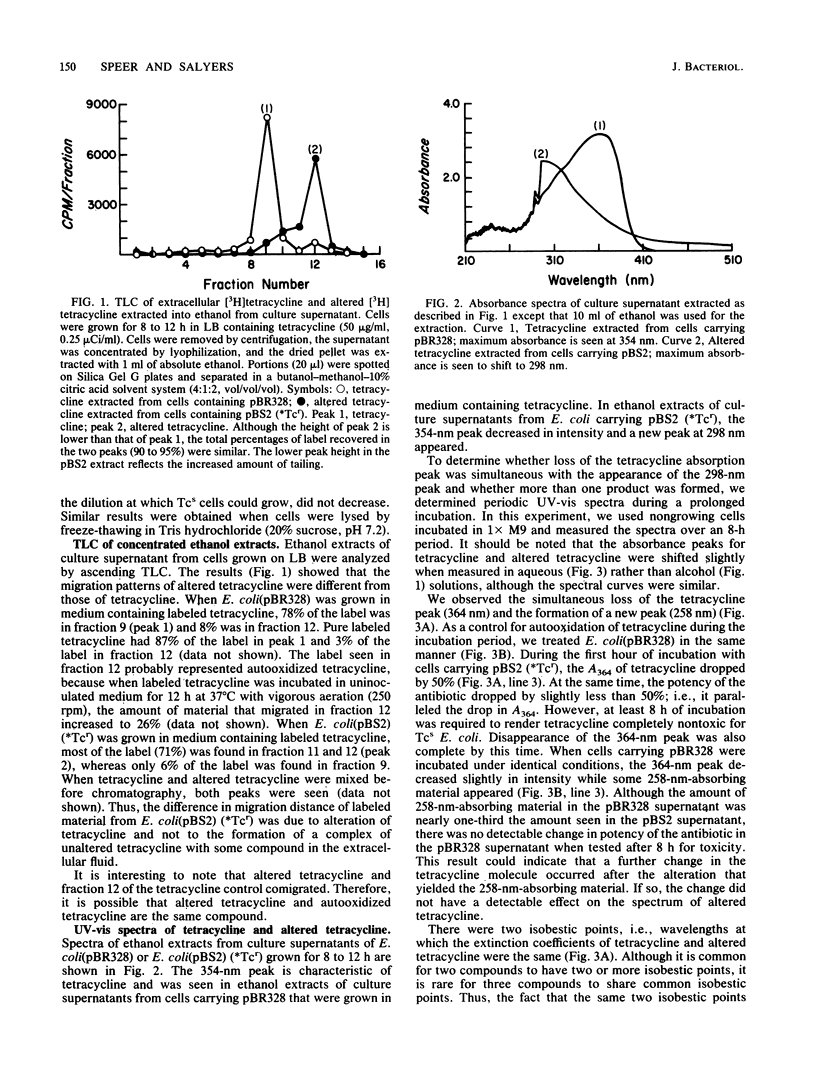
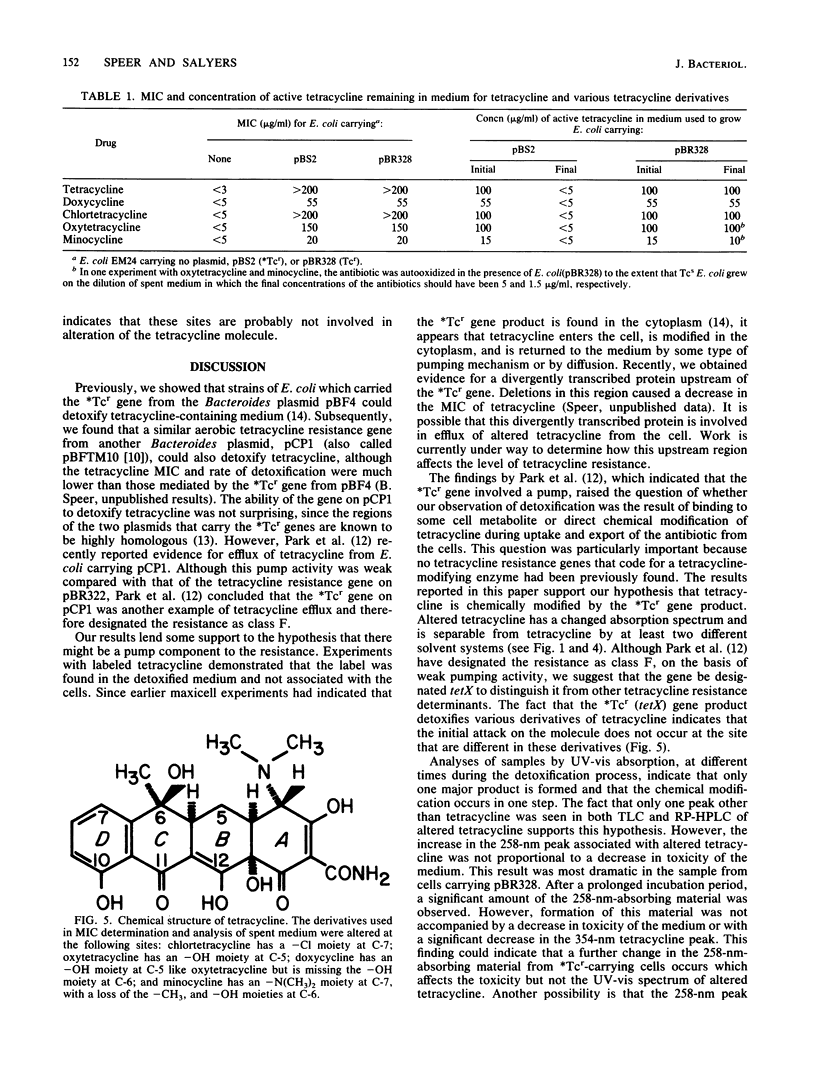
Abstract
A tetracycline resistance gene that was found originally on the Bacteroides plasmid pBF4 confers resistance on Escherichia coli but only when cells are growing aerobically. When E. coli EM24 carrying this aerobic tetracycline resistance (*Tcr) gene is grown in medium containing tetracycline, the resulting spent medium is no longer toxic to tetracycline-sensitive (Tcs) E. coli EM24 (B.S. Speer and A.A. Salyers, J. Bacteriol. 170: 1423-1429, 1988). To determine whether the *Tcr gene product modified tetracycline, we characterized the material resulting from incubation of E. coli (*Tcr) with tetracycline. When [7-3H(N)]tetracycline was added to cultures of E. coli (*Tcr), at least 90% of the label was recovered in the extracellular fluid. Therefore, tetracycline was not being sequestered by the cells. The labeled material behaved similarly to tetracycline with respect to solubility in various organic solvents. However, the UV-visible light spectrum had a single peak at 258 nm, whereas the tetracycline spectrum had a peak at 364 nm. The labeled material also had a faster migration rate than did tetracycline on thin-layer plates in a solvent system of butanol-methanol-10% citric acid (4:1:2, vol/vol/vol) and was separable from tetracycline by reverse-phase high-pressure liquid chromatography, using an acetronitrile-0.1% trifluoroacetic acid solvent system. These results demonstrate that the *Tcr gene product chemically modifies tetracycline. The *Tcr gene is the first example of a chemically modifying tetracycline resistance mechanism.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Burdett V., Inamine J., Rajagopalan S. Heterogeneity of tetracycline resistance determinants in Streptococcus. J Bacteriol. 1982 Mar;149(3):995–1004. doi: 10.1128/jb.149.3.995-1004.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdett V. Streptococcal tetracycline resistance mediated at the level of protein synthesis. J Bacteriol. 1986 Feb;165(2):564–569. doi: 10.1128/jb.165.2.564-569.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra I., Howe T. G. Bacterial resistance to the tetracyclines. Microbiol Rev. 1978 Dec;42(4):707–724. doi: 10.1128/mr.42.4.707-724.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covarrubias L., Cervantes L., Covarrubias A., Soberón X., Vichido I., Blanco A., Kupersztoch-Portnoy Y. M., Bolivar F. Construction and characterization of new cloning vehicles. V. Mobilization and coding properties of pBR322 and several deletion derivatives including pBR327 and pBR328. Gene. 1981 Jan-Feb;13(1):25–35. doi: 10.1016/0378-1119(81)90040-8. [DOI] [PubMed] [Google Scholar]
- Foster T. J. Plasmid-determined resistance to antimicrobial drugs and toxic metal ions in bacteria. Microbiol Rev. 1983 Sep;47(3):361–409. doi: 10.1128/mr.47.3.361-409.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiney D. G., Jr, Hasegawa P., Davis C. E. Expression in Escherichia coli of cryptic tetracycline resistance genes from bacteroides R plasmids. Plasmid. 1984 May;11(3):248–252. doi: 10.1016/0147-619x(84)90031-3. [DOI] [PubMed] [Google Scholar]
- Hasan T., Cooperman B. S. Reversed-phase high-performance liquid chromatographic separations of tetracycline derivatives using volatile mobile phases. J Chromatogr. 1985 Mar 15;321(2):462–466. doi: 10.1016/s0021-9673(01)90465-0. [DOI] [PubMed] [Google Scholar]
- Matthews B. G., Guiney D. G. Characterization and mapping of regions encoding clindamycin resistance, tetracycline resistance, and a replication function on the Bacteroides R plasmid pCP1. J Bacteriol. 1986 Aug;167(2):517–521. doi: 10.1128/jb.167.2.517-521.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park B. H., Hendricks M., Malamy M. H., Tally F. P., Levy S. B. Cryptic tetracycline resistance determinant (class F) from Bacteroides fragilis mediates resistance in Escherichia coli by actively reducing tetracycline accumulation. Antimicrob Agents Chemother. 1987 Nov;31(11):1739–1743. doi: 10.1128/aac.31.11.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. J., Gonda M. A. Comparison of the transposon-like structures encoding clindamycin resistance in Bacteroides R-plasmids. Plasmid. 1985 May;13(3):182–192. doi: 10.1016/0147-619x(85)90041-1. [DOI] [PubMed] [Google Scholar]
- Speer B. S., Salyers A. A. Characterization of a novel tetracycline resistance that functions only in aerobically grown Escherichia coli. J Bacteriol. 1988 Apr;170(4):1423–1429. doi: 10.1128/jb.170.4.1423-1429.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]


