Abstract
In the absence of the telomerase, telomeres undergo progressive shortening and are ultimately recruited into end-to-end chromosome fusions via the non-homologous end joining (NHEJ) double-strand break repair pathway. Previously, we showed that fusion of critically shortened telomeres in Arabidopsis proceeds with approximately the same efficiency in the presence or absence of KU70, a key component of NHEJ. Here we report that DNA ligase IV (LIG4) is also not essential for telomere joining. We observed only a modest decrease (3-fold) in the frequency of chromosome fusions in triple tert ku70 lig4 mutants versus tert ku70 or tert. Sequence analysis revealed that, relative to tert ku70, chromosome fusion junctions in tert ku70 lig4 mutants contained less microhomology and less telomeric DNA. These findings argue that the KU-LIG4 independent end-joining pathway is less efficient and mechanistically distinct from KU-independent NHEJ. Strikingly, in all the genetic backgrounds we tested, chromosome fusions are initiated when the shortest telomere in the population reaches ∼1 kb, implying that this size represents a critical threshold that heralds a detrimental structural transition. These data reveal the transitory nature of telomere stability, and the robust and flexible nature of DNA repair mechanisms elicited by telomere dysfunction.
INTRODUCTION
A primary function for the telomere is to confer a protective end structure that prevents natural chromosome ends from being inappropriately recognized as double-strand breaks (DSBs). This is accomplished by the specialized architecture at the chromosome terminus. In most eukaryotes, telomeres are comprised of stretches of TG-rich repeated DNA sequences that terminate in a single-strand overhang (G-overhang), and are bound by double- and single-strand specific telomere proteins (1). For added protection, telomeres can assemble into a higher order t-loop configuration that apparently unfolds in S phase to allow telomerase access for telomeric DNA synthesis.
Telomere function can be disrupted by prolonged telomerase inactivation or by perturbation of telomere-binding proteins. In such settings, the telomere triggers a DNA damage response and is processed as a DSB (2). One outcome is the fusion of aberrant telomeres end-to-end through the non-homologous end-joining (NHEJ) repair pathway (2). Telomere fusion leads to the formation of dicentric chromosomes that in anaphase form bridges only to be broken when chromosomes are segregated. The new DSBs induce breakage–fusion–bridge cycles, resulting in chromosome rearrangements that severely compromise genome stability.
The core components of the NHEJ machinery include LIG4/XRCC4 and the KU70/80 heterodimer (3). The KU complex acts to juxtapose two DSBs in alignment, while LIG4 and its stabilizing partner, XRCC4, ligate the two ends. In budding and fission yeast, the absence of KU and/or LIG4 leads to severe defects in end-joining; a 10–400-fold decrease in NHEJ has been reported (4–7). Likewise, mammalian cells deficient in KU and/or LIG4 display up to a 10-fold decrease in NHEJ (8–12). In both yeast and mammals, KU-independent end-joining pathways have been described, which rely on microhomology for alignment of the termini. In yeast, microhomology-mediated end-joining (MMEJ) is driven by the MRX complex (13). In vitro studies in human cells also demonstrate that the MRN complex utilizes microhomology to mediate end-joining (14), but it is currently unclear whether this pathway operates in vivo Recent studies also implicate PARP1 and XRCC1/DNA ligase 3 (LIG3) in the repair of DSBs in mammalian cells lacking KU and LIG4 (15–17).
In addition to its role in DSB repair, KU localizes to telomeres where it functions both in telomere length maintenance and chromosome end protection (2). Notably, the loss of KU in vertebrates and fission yeast results in an increased incidence of end-to-end chromosome fusions (18–22). In this respect it is paradoxical that KU, a key component of the NHEJ machinery, actively blocks telomere fusion. How KU can provide stability to chromosome ends without engaging NHEJ is unclear, but one possibility is that telomere binding proteins occlude active sites on KU essential to DNA repair (23).
As in DSB repair, fusion of dysfunctional telomeres can be mediated by canonical NHEJ as well as alternate end-joining pathways. In budding yeast, LIG4 is required for joining dysfunctional telomeres to internal DSBs (24). Similarly, studies in both mammalian cells and fission yeast indicate that the fusion of dysfunctional telomeres is dependent on LIG4 (25,26). Notably, in the former study where telomere de-protection was induced by the loss of an essential telomere-binding protein, telomeres remained in an open, stabile configuration with intact G-overhangs, even though the ends were recognized as DNA damage (25). In contrast, dysfunctional telomeres that arise in fission yeast and mammalian cells as a consequence of a long-term telomerase deficiency fuse efficiently in the absence of LIG4 (27,4). Hence, the context in which the telomere is de-protected may influence its processing by DNA repair machinery.
Due to its genetic tractability and high tolerance for genome instability, Arabidopsis is a useful model for studying the consequences of telomere dysfunction. Wild-type Arabidopsis telomeres range in size from 2 to 5 kb (28), but in mutants lacking the telomerase catalytic subunit, TERT, telomeres shorten by ∼200–500 bp per generation, ultimately triggering the formation of abundant end-to-end chromosome fusions (29,30). In the terminal generation of the mutants, where plants were sterile and unable to propagate to the next generation, the shortest functional telomere bearing an intact G-overhang is only ∼300 bp (31).
We previously showed that critically shortened telomeres fuse with approximately the same efficiency in the presence or absence of KU (32), arguing that plants employ highly flexible pathways for NHEJ. Since Arabidopsis telomeres are abutted by unique subtelomeric sequences on most chromosome arms, it is feasible to investigate mechanisms of chromosome end-joining at the molecular level using PCR. Chromosome fusion junctions arising in tert mutants display canonical NHEJ signatures, including the insertion of filler DNA, microhomology and nucleotide deletions (31). The average amount of telomere DNA captured in chromosome fusion junctions is 270 bp, consistent with the length of the shortest functional telomere. While the overall architecture of fusion junctions is similar in tert ku70 mutants, insertion of filler DNA is reduced and the majority of fusions possess microhomology, suggesting a microhomology-dependent back-up pathway. Consistent with this prediction, chromosome fusions that arise in triple tert ku70 mre11 mutants display decreased microhomology and increased insertions (31), strongly implicating the MRX/N complex in a backup pathway for chromosome end-joining.
Here we examine the role of LIG4 in telomere fusion. We find that critically shortened telomeres can still fuse in plants lacking LIG4 and KU, with only a modest decrease in frequency. However, the chromosome fusion junctions display different sequence signatures than in tert ku70 mutants, and the termini exhibit evidence for increased nucleolytic processing prior to fusion, arguing that the KU-LIG4 independent repair pathway is both mechanistically distinct, and less efficient than KU-independent NHEJ. Finally, we report a novel and critical size threshold for Arabidopsis telomeres that heralds the onset of chromosome end de-protection.
MATERIALS AND METHODS
Plant growth and MMS treatment
Arabidopsis thaliana were grown and DNA was extracted as described (33) with an exception noted below. For methyl methanesulfonate sensitivity (MMS), seeds of wild-type, ku70 and lig4-4 were sterilized in 50% bleach and plated on solid 0.5 BM media (34). Four-day-old seedlings were transferred to separate wells of a 24-well plate containing liquid 0.5 BM medium containing 0, 0.006, 0.008 or 0.01% MMS (Aldrich) and incubated in a shaker with constant light. Seedlings were scored after 3 weeks.
Generation of lig4 mutants and complementation of MMS sensitivity in lig4-4
Arabidopsis thaliana plants with a T-DNA insertion in AtLIG4 were obtained from the Salk collection (line 04427) (35). Heterozygous plants were identified using PCR with primers Lig4–8 (5′ GTGATTTGAAACTAGTCTGTG 3′), Lig4–9 (5′ CAGCAAACCGATTCAGAGATG 3′) and LbA–1 (5′ TGGTTCACGTAGTGGGCCATCG 3′). Plants heterozygous for T-DNA insertion in KU70 and TERT (29,33) were crossed to heterozygous lig4-4. PCR was used to identify a triple heterozygous plant in F1, and this plant was self-pollinated to produce a segregating F2 population (Figure 2A). All single, double and triple mutants along with wild-type plants were identified by PCR from this population. The genomic LIG4 coding sequence (Genbank accession number: AB023042) was PCR-amplified using DNA from wild-type Arabidopsis plants and placed under direction of the cauliflower mosaic virus promoter and the octopine synthase transcriptional terminator and cloned into a vector (36) for use in agrobacterium-mediated plant transformation (37). The MMS sensitivity of the lig4-4 mutation was complemented with this genomic construct (data not shown). The experiment shown in the last panel of Figure 2C was performed with DNA from plants derived from a cross between tert and lig4-1 mutants (37). The lig4-1 allele produces a LIG4 transcript, 3′ of the T-DNA insertion site presumably from a cryptic promoter within the T-DNA. However, due to the presence of stop codons within the inserted T-DNA construct, this allele likely produces a non-functional protein. In support, lig4-1 mutants exhibit sensitivity to DNA damage, expected from the loss of a DNA repair protein.
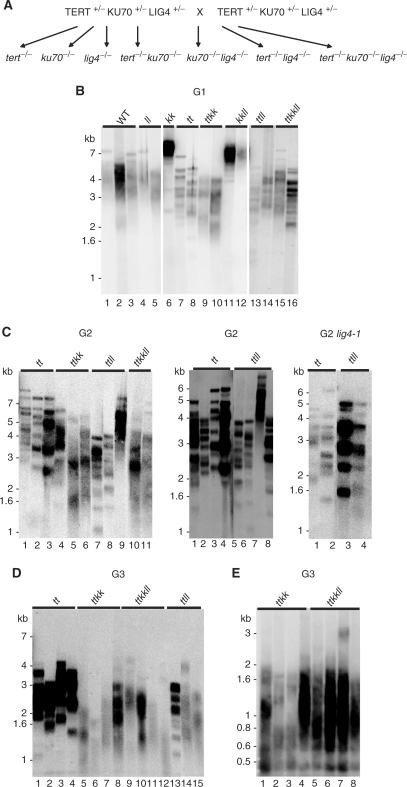
Figure 2.
Characterization of telomere length in tert ku70 lig4 mutants. (A) Schematic representation of genetic crosses showing the segregation of various lig4 mutants resulting from self-pollination of a triple heterozygous TERT+/− KU70+/− LIG4+/− parent. (B–D) TRF analysis of generation 1 (G1) (B), G2 (C), and G3 (D) mutant combinations arising from segregation of the triple heterozygote shown in (A). The right panel in Figure C shows TRF data for a tert mutants carrying the lig4-1 allele (37). (E) TRF analysis of G3 tert ku70 and tert ku70 lig4 mutants that have reached the terminal phenotype. Genotype abbreviations are as follows: wild-type (WT), lig4 (ll), ku70 (kk), tert (tt), ku70 lig4 (kkll), tert lig4 (ttll) and tert ku70 lig4 (ttkkll). The differences in hybridization signals reflect slight variations in the amount of DNA loaded in each lane.
Nucleic acid extraction, RT–PCR and telomere fusion PCR
DNA was extracted using the CTAB method (38). For RT–PCR, total RNA was extracted from flowers using the TriReagent solution (Sigma, St Louis, MO). RT was performed using 1 μg of total RNA with SuperScript III (Invitrogen) and oligo dT at 55°C. The following primer pairs were used: Lig4–1 (5′ ATGACGGAGGAGATCAAATTCAGCG 3′) with Lig4–2 (5′ TGACCCACTTCATCTCCTGAGC 3′), Lig4–8 with Lig4–9 (both sequences described above), and Lig4–5 (5′ GGGAACCTGGAGATCGTAGTGG 3′) with Lig4–6 (5′ TGC CCTTGATATCCGATACATCAG 3′). Telomere fusion PCR was performed as previously described (31).
TRF, subtelomere analysis and Primer Extension Telomere Repeat Amplification (PETRA)
For TRFs and subtelomere analysis, ∼1 µg of DNA was digested with 30 U of the restriction endonuclease TruII overnight at 65°C. DNA was recovered by ethanol precipitation, suspended in water and loaded into a 0.8% agarose gel run at 50 V for ∼16 h. The gel was transferred onto a nylon membrane (Hybond), hybridized with a [γ32P]ATP end-labeled (T3AG3)4 oligonucleotide in a buffer containing 0.25 M sodium phosphate buffer (pH 7.5), 7% SDS and 1 mg/ml BSA. Hybridization signals were detected using a STORM PhosphorImager (Molecular Dynamics) and the data were analyzed using IMAGEQUANT software (Molecular Dynamics). Specific subtelomeres were assessed in the same manner except that the DNA was digested overnight at 37°C with 30 U of PveII and SpeI in order to release intact subtelomeres (and their respective telomeres) from the bulk of genomic DNA. The probes used in the detection of the subtelomeres 1L, 2R and 5L are described previously (28). The length of specific telomere tracts was also determined using PETRA as described previously (38) except that in most cases whole plant tissue was used.
In-gel hybridization
In-gel hybridization was performed essentially as described (39) with some modifications. Approximately 300 mg of plant tissue was extracted using a GE DNA extraction kit (product code 27-5237-01) as per the manufacturer's instructions, except that the tissue was incubated at 37°C. Extracted DNA was incubated with SSB protein (Promega) to a final concentration of 2 µg/ml, to protect the single-stranded G-overhang from degradation. The DNA was then subjected to digestion using 30 U of each of the restriction endonucleases, HaeIII and HinfI, overnight at 37°C. The gel was hybridized with a [γ32P]ATP end-labeled (TA3C3)3 oligonucleotide. To ensure the signal obtained was from a single-stranded 3′ G-overhang, DNA was either mock or treated with T4 DNA polymerase (utilizing its 5′ to 3′ exonuclease activity). As expected, a loss of signal was observed in the samples treated with 24 U of T4 DNA polymerase (Figure 5B). Agarose gels were denatured and re-hybridized using the same probe. The single-strand G-overhang signals were obtained by calculating the volume of the signal in each lane and then normalizing the signal using the hybridization signal obtained from the denatured gel. The single-strand G-overhang signal obtained from wild-type was set to one and each sample was normalized to this value.
Figure 5.
Subtelomere and G-overhang analysis in tert ku70 lig4 mutants. (A) Subtelomere TRF analysis using probes specific for 1L (left blot), 2R (middle blot) and 5L (right blot). DNA was analyzed from tert, line 12 (lanes 1–3), tert ku70, line 12 (lanes 4–7) and tert ku70 lig4, line 9 (lanes 8–10). (B) In-gel hybridization of DNA isolated from wild-type plants under native (left panel) and denaturing conditions (right panel). Lane 2 from each gel shows DNA subjected to T4 DNA polymerase (Exo) treatment, demonstrating the signal is dependent on the G-overhang. (C) In-gel hybridization of DNA isolated from wild-type and mutant plants. The wild-type control is shown in lane 1. Duplicate DNA obtained from line 3 for tert ku70 are shown in lanes 2–5 and from two separate pools from three separate plants from line1 for tert ku70 lig4 are shown in lanes 6–9. The hybridization signal for each lane was normalized to the wild-type sample and average fold increase over the wild-type signal is reported. The denatured gel showing bulk telomeres is shown on the right. All gels were hybridized with a [γ32P] ATP radiolabeled (TA3C3)3 telomere repeat probe. Genotype abbreviations used are as stated in Figure 2.
Cytogenetic analysis
Mitotic anaphases were obtained from pistils of unopened floral buds as described in (30) but 8-hydroxyquinoline was omitted. Squashes were analyzed with a Zeiss epifluorescence microscope. The anaphase bridges were scored as a percentage of total anaphases (Table 1).
Table 1.
Summary of cytogenetic analysis
| Genotype | Anaphase bridges | Total anaphases | Percent bridgesa |
|---|---|---|---|
| Generation 3 | |||
| tert ku70 (line 3) | 108 | 1641 | 6.6 |
| tert ku70 (line 12)b | 237 | 1060 | 22.3 |
| tert ku70 lig4 (line1) | 5 | 1714 | 0.3 |
| tert ku70 lig4 (line 9)b | 46 | 762 | 6 |
| Generation 4 | |||
| tert ku70 (line 3) | 340 | 1195 | 28.4 |
| tert ku70 lig4 (line1) | 139 | 1354 | 10.3 |
aFor each generation, genoptype and line, the total number of anaphase bridges was divided by the total anaphases observed.
bIndicates the generation and line where the plants were terminal.
Statistical analysis
Statistical analysis was performed using a Student's t-test and a P-value <0.05 was considered significant.
RESULTS
LIG4 is not required for telomere length homeostasis in Arabidopsis
We characterized a SALK T-DNA insertion line for LIG4, which we term lig4-4, where the insertion lies in the 6th exon (Figure 1A). To determine whether the LIG4 gene was still functional, RT–PCR was conducted to monitor the level of LIG4 mRNA (Figure 1B). Although we detected evidence for transcripts both upstream and downstream of the T-DNA insertion, no RT–PCR products were observed with primers flanking the T-DNA junction (Figure 1B). Since this region includes the conserved active site lysine required for catalytic activity, the data argue that LIG4 is inactive. Consistent with this conclusion, treatment of lig4-4 mutants with 0.01% of the DSB-inducing agent MMS led to growth arrest, while wild-type plants thrived under the same conditions (data not shown). We verified that the DNA repair defect was linked to the LIG4 gene by transforming a wild-type genomic copy of LIG4 into lig4-4. MMS hypersensitivity of the transformants was abolished (data not shown). We conclude that the lig4-4 is a null allele of LIG4.
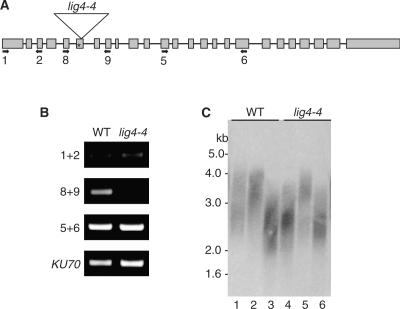
Figure 1.
Characterization of the lig4-4 mutant. (A) Schematic representation of the Arabidopsis LIG4 gene showing positions of 25 exons (rectangles); the T-DNA is inserted in the 6th exon (triangle). The active site lysine is indicated by an asterisk. Primer positions are denoted by arrows. (B) RT–PCR analysis using primer combinations shown in (A). There is no transcript detected with primers that flank the T-DNA junction using primers 8 and 9. Products downstream of the T-DNA insertion likely are derived from a cryptic promoter in the T-DNA construct (49). (C) Terminal Restriction Fragment (TRF) analysis of lig4-4 mutants and wild-type plants segregated from LIG4-4+/− plants. Although telomeres in lig4-4 mutants (lanes 4–6) appear more variable in length than in wild-type (lanes 1–3), they fall within wild-type range.
As expected from previous analysis of other lig4 Arabidopsis lines (37,40), lig4-4 mutants were viable and showed no developmental defects under normal growth conditions (data not shown). To determine if LIG4 contributes to telomere maintenance in Arabidopsis, terminal restriction fragment (TRF) analysis was performed on DNA isolated from lig4-4. Although individual telomere lengths varied slightly, there was no significant difference in the telomere tracts of mutants versus their wild-type siblings. All telomeres migrated in the 2–5-kb range (Figure 1C, compare lanes 1–3 with lanes 4–6). In agreement with previous studies in yeast and mammals (4,7,18), and a previous report for Arabidopsis (40), we conclude that LIG4 is not required for telomere length homeostasis. Furthermore, consistent with their wild-type phenotype, lig4-4 mutants do not display evidence for genome instability (see subsequently).
Disruption of LIG4 does not accelerate the onset of the terminal phenotype in plants with critically shortened telomeres
To investigate the role of LIG4 in promoting telomere fusions, the lig4-4 allele was crossed into a genetic background where telomeres are rapidly shortening. Telomeric DNA is lost 2- to 3-fold faster in plants null for both TERT and KU70, and mutants reach the terminal phenotype as early as the third generation (G3) of the mutant (32). In contrast, tert mutants typically survive until at least G8 (30). To accelerate our analysis of LIG4, we generated a plant heterozygous for TERT, KU70 and LIG4 and allowed it to self-pollinate to create a triple tert ku70 lig4 mutant (Figure 2A). From this cross we isolated all combinations of single, double and triple mutants and evaluated bulk telomere length from each genotype combination by TRF. Figure 2 shows the results of this analysis for successive generations of mutant lines arising from the triple heterozygote. As expected, telomere shortening was observed in all the plants lacking a functional TERT gene (Figure 2B, lanes 7–10, 13–16), and was accelerated in double tert ku70 mutants (Figure 2B, lanes 9 and 10). Furthermore, as expected, ku70 mutants displayed greatly extended telomeres (Figure 2B, lane 6), consistent with our previous observation that KU70 acts as a negative regulator for telomere length (33).
Although there were some variations in telomere length among sibling plants, inactivation of LIG4 in combination with a KU70 or TERT deficiency gave rise to telomere phenotypes that were similar to those associated with single mutants in G1 (Figure 2B). As expected, LIG4 deficiency neither abolished telomere extension in ku70 mutants, nor did telomere length vary significantly in tert lig4 versus tert or in tert ku70 versus tert ku70 lig4. However, in G2 and G3, the rate of telomere shortening appeared to be slightly faster in several tert lig4 mutants relative to tert (Figure 2C, left panel, compare lanes 1–3 with 7 and 8; middle panel, compare lanes 1–4 with 5, 6 and 8; Figure 2D compare lanes 1–4 with 13–15). Furthermore, PETRA analysis of individual telomeres in later generation tert lig4 mutants showed telomeres that were somewhat shorter than in tert (data not shown). Confounding these results, however, was a three-generational analysis of tert lig4 mutants (G1–G3) derived from a second LIG4 mutant allele [lig4-1; (37)]. Similar to lig4-4, lig4-1 appears to be a null allele displaying the sensitivity to DNA damage (37). These double mutants did not reveal evidence for accelerated telomere shortening relative to their tert counterparts (Figure 2C, right panel; data not shown). Therefore, our genetic analysis indicates that LIG4 may make a modest contribution to telomere length regulation in the context of a telomerase deficiency, but further studies are needed to address this possibility.
Notably, a generational analysis failed to reveal differences in the rate of telomere shortening in triple tert ku70 lig4 mutants versus tert ku70 (Figure 2E). Consistent with similar rates of telomere attrition, both tert ku70 and tert ku70 lig4 reached the terminal phenotype as early as G3 or G4. To correlate this phenotype with telomere length, we sought to determine the minimal functional telomere length in the most severely affected, developmentally arrested plants using PETRA (31). PETRA is a sensitive, PCR-based technique that targets specific telomere arms using a primer directed at a unique subtelomeric sequence and a primer that anneals to the 3′ G-overhang. This approach allows us to accurately measure telomere length on 7/10 chromosome ends (31). We define a functional telomere as harboring an intact G-overhang, and hence the minimal functional length corresponds to the shortest PETRA products generated. The shortest telomeres that we could detect in a pool of terminal tert ku70 mutants was 360 bp (31) and in the two fourth generation tert ku70 lig4 mutants were 320 and 450 bp (Figure 3A and Supplementary Table 2). These data support our conclusion that 300 bp represents the minimal functional length for Arabidopsis telomeres, below which the telomere is unable to maintain a G-overhang and block end-to-end fusion. Thus, the failure of tert ku70 lig4 to proliferate beyond G3 or G4 is likely due to the accumulation of non-functional telomere caps.
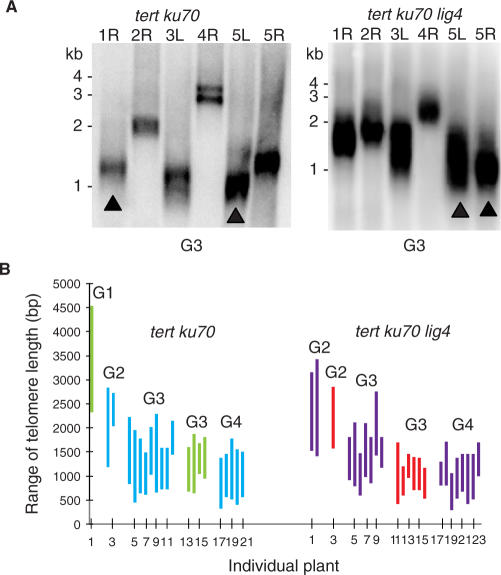
Figure 3.
Analysis of telomeres in tert ku70 lig4 mutants. (A) Representative PETRA data for G3 tert ku70 and tert ku70 lig4 mutants. Closed arrows indicate the two shortest telomere tracts, determined by subtracting the length determined by PETRA from the relative position of the chromosome-specific primer on the subtelomere DNA target. (B) Summary of PETRA results for individual plants from tert ku70 and tert ku70 lig4 lines. Vertical lines represent that range of telomere lengths (shortest to longest) detected in each plant. Blue and green lines represent tert ku70 line 3 and line 12; purple and red lines denote tert ku70 lig4 lines 1 and 9. The generation for each line is denoted above each group of vertical lines.
LIG4 is not essential for the fusion of critically shortened telomeres
We asked whether LIG4 is necessary to fuse critically shortened telomeres by comparing the efficiency of chromosome end-joining events in tert ku70 versus tert ku70 lig4 plants. Telomere fusion was evaluated initially using conventional cytogenetics to detect bridged chromosomes in anaphase. Two independent lines from tert, tert ku70, tert lig4 and tert ku70 lig4 mutants were analyzed for three consecutive generations (Table 1). As expected, no anaphase bridges were observed in G1 or G2 of any of the mutants (data not shown). However, beginning in G3 anaphase bridges were observed in both lines of tert ku70 and tert ku70 lig4 mutants. Thus, LIG4 is not required for chromosome fusions in cells with critically shortened telomeres.
For plants that had not reached the terminal phenotype, and which had comparable telomere lengths, the percentage of anaphase bridges was not statistically different. For example, the percentage of anaphase bridges in G3 tert ku70 line 3 was 6.6% and in G4 tert ku70 lig4 line 1 was 10.3% (Table 1). However, comparison of chromosome fusion rates in terminal generation mutants revealed that the efficiency of end-joining was modestly reduced in the absence of LIG4 (Table 1). In G3 tert ku70 lig4 (line 9) 6% of the anaphase showed bridges, while in G3 tert ku70 (line 12) 22.3% of the anaphases harbored bridges. This difference is statistically significant (P = 0.04). A similar trend was observed with two other lines in G4; the number of anaphase bridges in tert ku70 lig4 line 3 was reduced relative to tert ku70 line 1 mutants (10.3% vs 28.4%; P = 0.002). Thus, the frequency of chromosome end-joining events appears to be decreased in the absence of LIG4 by a factor of ∼3-fold.
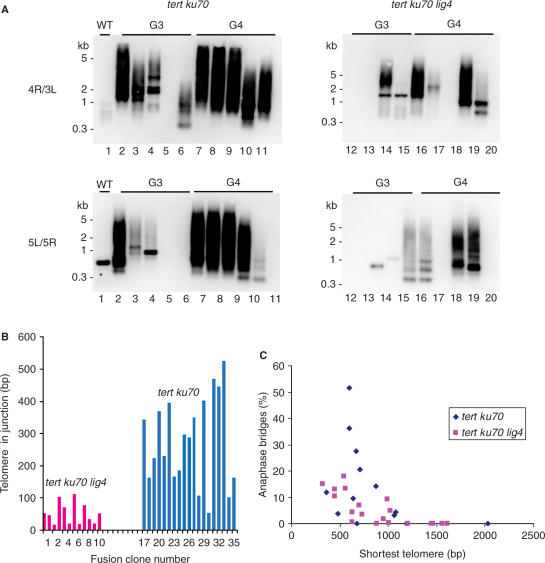
To verify that the anaphase bridges we observed reflect telomere fusions, we analyzed these same samples using telomere fusion PCR with primers directed outward from the right arm of chromosome 1 (1R) and the left arm of chromosome 3 (3L) (31). Southern blot analysis of the PCR products using a telomeric DNA probe gave a signal for tert ku70 lig4 reactions, although it was reduced compared with tert ku70 (Figure 4A). Notably, results from telomere fusion PCR correlated well with the reduced frequency of anaphase bridges for the majority (75%) of tert ku70 lig4 samples tested (Table 1 and Supplementary Table 2).
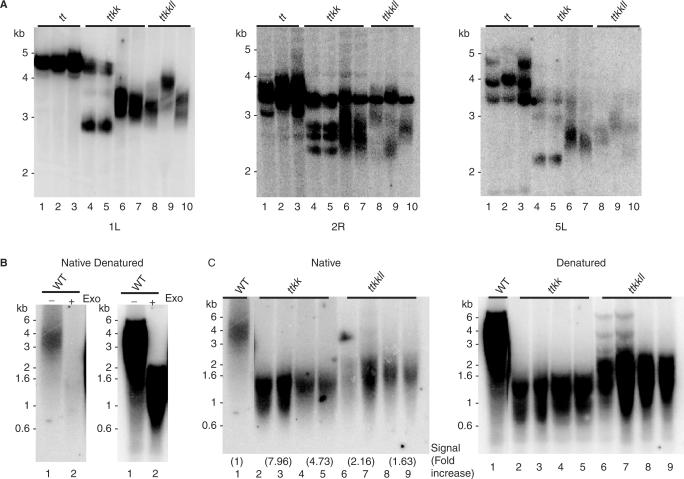
Figure 4.
Telomere length and the onset of end-to-end fusions in lig4 mutants. (A) Results are shown from telomere fusion PCR performed on individual wild-type and G3 and G4 tert ku70 and tert ku70 lig4 mutants. The primers used for PCR are indicated on the left. Blots were hybridized with a radiolabeled telomere repeat probe. Non-specific bands are occasionally observed (*) in this assay, but can be discerned by sequencing the cloned product. (B) Graph illustrates the amount of telomeric DNA in fusion junctions arising from tert ku70 lig4 (pink bars) and tert ku70 mutants (blue bars). Data were obtained from sequence analysis of telomere fusion PCR products derived from tert ku70 lig4 mutants (this study) and from tert ku70 mutants (31). (C) Graph illustrates the relationship between shortest telomere and the frequency of anaphase bridges in G1–G4 of tert ku70 and tert ku70 lig4.
One explanation for the decreased amount of telomere fusion PCR products is that chromosome ends in the triple mutants were subjected to extensive nucleolytic degradation into the subtelomeric region prior to fusion, which could eliminate primer-binding sites. However, when PCR was performed with primers directed at slightly more internal sites on the chromosome, product abundance was not increased. An alternative possibility is that the telomere arms we targeted were not critically shortened, and hence would not be recruited into fusions. To address this issue, we determined the length of individual telomere tracts using PETRA. PETRA showed that the 5L and 5R telomeres were the very shortest in tert ku70 lig4 mutants (Figure 3A and Supplementary Table 2). When telomere fusion PCR was repeated using 5L and 5R primers, additional weak products were evident from several triple mutants after hybridization with a telomere repeat probe (Figure 4A). Hybridization of the blots with a probe directed to the 5L or 5R subtelomere also yielded only a faint signal (data not shown). These data support the conclusion that the frequency of chromosome end-joining reactions is reduced in the absence of LIG4.
Chromosome fusion junctions formed in the absence of KU70 and LIG4 display unique sequence signatures
In accordance with their reduced abundance, telomere fusion PCR products from triple mutants were difficult to clone. Nevertheless, sequence analysis was performed on 11 independent clones. We noted several interesting distinctions in the structure of fusion junctions formed in the presence and absence of LIG4. First, although the use of microhomology (defined by at least one nucleotide overlap) was prevalent in tert ku70 lig4 mutants, the amount of base pair overlap captured in the junctions was reduced in these mutants relative to tert ku70 (avg = 2 versus avg = 4.6 perfect nucleotide overlap, respectively; P = 0.02). Although the number of clones analyzed is small, these data suggest that end-joining reactions in the absence of both KU and LIG4 are mechanistically distinct from the KU-independent pathway. Second, critically shortened telomeres were more prone to nuclease attack in tert ku70 lig4 than in tert ku70. We observed a substantial decrease in the number of chromosome fusions that involved the direct joining of two telomere tracts. In tert ku70 mutants, 43% of the junctions reflected telomere-telomere joining, while this was true for only 18% of junctions analyzed from tert ku70 lig4 mutants [Table 2 and Supplementary Table 3; (31)]. In this regard, the chromosome fusion events in the triple mutant more closely resembled tert mutants, where only 11% of the junctions analyzed involved direct telomere-telomere joining (31). In both tert and tert ku70 lig4 mutants, the large majority of fusions (78 and 73%, respectively) corresponded to telomere–subtelomere joining events, in which one chromosome end had lost all of its telomeric DNA prior to fusion.
Table 2.
Characterization of chromosome fusion junctions in tert ku70 lig4 mutants
| tert ku70 n = 31b | tert ku70 lig4 n = 11 | |
|---|---|---|
| Type of fusiona | ||
| Telomere–telomere | 43% | 18% |
| Telomere–subtelomere | 51% | 73% |
| Subtelomere–subtelomere | 5% | 9% |
| Complex | 1% | 0% |
| Features of fusion junctions | ||
| Deletion of subtelomere sequence | 220 bp | 220 bp |
| Telomeric repeat retained at fusion site | 270 bp | 50 bp |
| Microhomology at fusion junctionc | 81% | 66% |
| Insertion of filler DNA at fusion junction | 10% | 0% |
Additional evidence that critically shortened telomeres are more susceptible to nuclease attack in lig4 mutants was revealed when the amount of telomeric DNA captured in the fusion junctions was considered. Five-fold less telomeric DNA was associated with fusions cloned from tert ku70 lig4 than from ku70 tert (average of 50 bp of telomeric DNA vs 270 bp, P = 0.01) (Figure 4B and Table 2).
Two additional assays were performed to investigate the extent to which chromosome ends in tert ku70 lig4 are subjected to nucleolytic attack (Figure 5). First, subtelomeric TRF analysis was employed. While the profile of bulk telomeres in terminal generation plants detected by standard TRF blots is indistinguishable in tert ku70 and tert ku70 lig4 mutants, a subset of chromosome ends could have suffered extensive nuclease attack. Although we did not have sufficient genomic DNA from terminal generation mutants to evaluate all the chromosome arms, analysis of the 1L, 2R and 5L telomeres indicated that at least these three termini have not been extensively degraded (Figure 5A). Second, we performed in-gel hybridization to monitor the status of the G-overhang in terminal generation tert ku70 lig4 mutants. Telomere uncapping in LIG4-deficient mammalian cells does not result in any loss of G-overhangs (25,41). In two separate experiments, we observed only a 2- to 3-fold decrease in the G-overhang signal for tert ku70 lig4 mutants relative to tert ku70 (Figure 5C; data not shown). This finding is not unexpected, as PETRA, which relies on the presence of an intact G-overhang, consistently yielded products in terminal tert ku70 lig4 mutants.
From these experiments we conclude that critically shortened telomeres in Arabidopsis tert ku70 lig4 mutants are subjected to increased, but not catastrophic nuclease attack, which likely reflects the decreased efficiency of end-joining catalyzed by a KU-LIG4 independent repair pathway.
Identification of a novel, critical size threshold for Arabidopsis telomeres
To gain a deeper understanding of the molecular trigger for telomere dysfunction and chromosome fusion in plants undergoing progressive telomere erosion, we monitored the dynamics of individual telomere tracts through consecutive generations of tert ku70 and tert ku70 lig4 mutants. PETRA analysis revealed that the absence of LIG4 did not affect the stochastic nature of which telomere is the shortest. For example, in one G4 tert ku70 lig4 mutant the shortest telomere was 5R (320 bp), while in another plant it was 4R (450 bp) (Supplementary Table 2). Similarly, the range of telomere lengths (i.e. the difference between the shortest and longest telomere in a given plant) was approximately the same in tert ku70 and tert ku70 lig4 mutants. The average range of telomere lengths for the two G3 tert ku70 lines was 990 bp and 930 bp in G4 (Figure 3B and Supplementary Table 1). For tert ku70 lig4 the range was 820 bp and 750 bp, respectively (Figure 3B and Supplementary Table 2).
Despite the stochastic nature of telomere dynamics in all the genetic backgrounds we examined (tert, tert ku70, tert ku70 lig4), the onset of telomere fusions strongly correlated with the presence of at least one telomere in the population that was 1 kb or below in length (Figure 4C and Supplementary Tables 1 and 2). In tert ku70 mutants 94% of the plants (15/16) shown to contain telomere fusions by our PCR assay harbored a telomere of ⩽1 kb (Supplementary Table 1). For tert ku70 lig4 mutants this value was 90% (9/10) (Supplementary Table 2). Similarly, when samples showing cytogenetic evidence for chromosome fusions from tert ku70 lig4 mutants are included with samples showing positive telomere fusion PCR data, 87% of the plants contain at least one telomere that is less than or equal to ∼1 kb in length (Supplementary Table 2). This correlation was also evident when data from both tert ku70 and tert ku70 lig4 are combined (Figure 4C). Thus, although bulk telomeres continue to shorten until they reach the minimal functional size of ∼300 bp, our data indicate 1 kb represents a critical length threshold. Telomeres that drop below this length have lost the ability to efficiently cap the chromosome terminus and begin to participate in end-joining reactions.
DISCUSSION
Here we exploit the genetic tractability of Arabidopsis to examine DNA repair pathways that are elicited in mutants experiencing progressive telomere erosion. Specifically, we evaluated the role of LIG4 in promoting the formation of end-to-end chromosome fusions. By employing plants that were doubly deficient in TERT and KU70, our analysis was expedited as telomeres shortened two to three times faster in this background than in single tert mutants (32). Furthermore, by inactivating both KU and LIG4, we severely crippled the conventional NHEJ machinery, allowing us to investigate potential backup mechanisms for DSB repair. As with yeast and mammals, we found that LIG4 does not make a significant contribution to telomere length maintenance in Arabidopsis (4,7,18,40). Interestingly, we observed a slight increase in the rate of telomere shortening in tert mutants carrying the lig4-4 allele, but not the lig4-1. However, since tert lig4-4 mutants are viable through at least G6 as are their tert siblings (Heacock,M. and Shippen,D., unpublished data), LIG4 appears to play only a minor role in telomere length maintenance when telomerase is inactivated. Consistent with this conclusion, we found that bulk telomeres shorten at the same rate in tert ku70 lig4 mutants as in tert ku70 mutants. In both settings plants typically reach the terminal phenotype in G3 or G4.
To evaluate the role of LIG4 in joining dysfunctional telomeres, we monitored genome integrity in plants with shortening telomeres using a combination of cytogenetic analysis and telomere fusion PCR. As with KU mutants (32), we found that chromosome fusions can be readily detected in plants lacking both LIG4 and KU. Since NHEJ is favored in higher eukaryotes by 1000-fold over homologous recombination (42), the other well-characterized DSB repair pathway, our findings reveal a second backup mechanism for DSB repair that does not involve canonical NHEJ. Studies in yeast and mammals support this conclusion. LIG4-independent fusion of critically shortened telomeres has been reported in fission yeast (4), and very recently in mammalian cells (27). Intriguingly, evidence for a backup pathway of DNA ligation in Arabidopsis has also been described in lig4 mutants during T-DNA integration, a process thought to be mediated by NHEJ (37,40). As in the current study, Friesner and Britt (37) report that T-DNA integration is reduced by 3-fold in the absence of LIG4.
Although little is known about this alternative mechanism for NHEJ, our data provide some insight into how it may engage dysfunctional telomeres. Several lines of evidence indicate that the backup pathway is less robust than KU-independent NHEJ. In addition to the 3-fold reduction in anaphase bridges, we found that telomere fusion PCR products were less abundant in tert ku70 lig4 relative to tert ku70, even when primers were used that specifically targeted the very shortest telomeres in the population. Furthermore, critically shortened telomeres in plants lacking both KU and LIG4 appear more vulnerable to nuclease attack than in plants deficient only in KU. Not only is the incidence of telomere–telomere fusions decreased, but also reduced is the length of the telomeric DNA tract captured in fusion junctions in tert ku70 lig4 mutants compared to all of the other genetic backgrounds we have examined, including tert, tert ku70, tert lig4 and tert ku70 mre11 (Figure 4B and Table 2) (31).
We hypothesize that the increased nucleolytic digestion of dysfunctional telomeres in tert ku70 lig4 mutants is a reflection of reduced efficiency of repair. We found no evidence for extensive degradation of individual chromosome ends. Preliminary data from four independent telomere fusion clones from G5 tert lig4 mutants revealed an average of 125 bp of telomeric DNA in the junctions (Heacock,M. and Shippen,D., unpublished data), more than twice the amount of telomeric DNA recovered from fusions in triple tert ku70 lig4 mutants. Thus, the presence of KU in tert lig4 mutants may limit nucleolytic digestion of dysfunctional telomeres (43). It is noteworthy that G-overhang signals in terminal tert ku70 lig4 mutants are reduced relative to tert ku70 double mutants. Hence, although critically shortened telomeres lose their ability to effectively block NHEJ, they largely retain their capacity to protect the chromosome terminus.
What is the mechanism for chromosome end-joining in the absence of KU and LIG4? Our sequence analysis reveals that chromosome fusion junctions in tert ku70 lig4 mutants utilize shorter tracts of microhomology; tert ku70 mutants employ twice the amount of perfect nucleotide overlap (31). Thus, end-joining by the LIG4-KU independent pathway appears to be mechanistically distinct from the KU-independent pathway. The MRN complex, which has been implicated in KU-independent fusion of critically shortened telomeres in Arabidopsis (31), is known to mediate end-joining primarily through the use of microhomology (13,14) in an alternative pathway only partially dependent on LIG4 (13). As we see only a small difference in the level of microhomology at fusion junctions, we cannot rule out the possibility that MRN joins dysfunctional telomeres in KU-LIG4 mutants. Alternatively, end-joining could be mediated through a PARP1/LIG3-dependent pathway akin to what has been described in mammals (15–17). Although there is a clear PARP1 ortholog in the Arabidopsis genome, LIG3 is absent. LIG1 could act in combination with PARP1 to join DNA ends, as in vitro studies indicate that the mammalian LIG1 can join double-strand DNA breaks (44). It is also possible that LIG6, a DNA ligase unique to Arabidopsis whose function is unknown, substitutes for LIG4 (45).
Whatever the mechanism of end-joining dysfunctional telomeres, the molecular triggers for this reaction appear to be remarkably consistent in the presence or absence of key components of the NHEJ machinery. Our data indicate that Arabidopsis telomeres undergo at least two distinct structural changes en route to complete dysfunction. By measuring individual telomere tracts in plants undergoing progressive telomere shortening, we discovered that chromosome fusions are first initiated when the shortest telomere in the population reaches a size of ∼1 kb. One caveat of our experiments is that the PETRA technique can currently measure telomere lengths on only 7/10 Arabidopsis chromosome ends. Thus, it is formally possible that telomeres shorter than 1 kb are required to trigger chromosome fusions. Nevertheless, our data argue strongly that telomeres breach a length threshold at or near 1 kb. At this point, telomere tracts may be unable to assume a stable t-loop configuration, perhaps due to the reduced occupancy of some essential telomere binding proteins. An alternative, but not mutually exclusive model is that the reduced occupancy of telomere proteins on the shortened telomere releases the constraints on the telomere-associated DNA damage proteins, allowing them to instigate a DNA damage response that activates repair. Since plants survive for multiple generations after the initial onset of telomere dysfunction and their telomeres continue to shorten, this length threshold does not automatically incite significant genome instability. We speculate that shortened, ‘meta-stable’ telomeres can assume an alternative structure that affords the terminus protection from a full-blown DNA damage response. Such a structure could be a simple fold-back conformation proposed for yeast telomeres (46–48). As telomere erosion continues, the incidence of chromosome fusions increases until the telomere tract reaches the minimal length of residual function (∼300 bp) (31). Below this second size threshold, all of the features that distinguish the telomere from a DSB are lost.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We are grateful to the Shippen lab for many helpful discussions, Dmitri Churikov and Carolyn Price for providing the in-gel hybridization protocol and Steve Heacock for software expertise. This study was supported by NIH (GM065383) to D.E.S and by NIEHS (5-32-ES07059) to J.D.F. Funding to pay the Open Access publication charges for this article was provided by Grant NIH GM065383.
Conflict of interest statement. None declared.
REFERENCES
- 1.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 2.Riha K, Heacock ML, Shippen DE. The role of the nonhomologous end-joining DNA double-strand break repair pathway in telomere biology. Annu. Rev. Genet. 2006;40:237–277. doi: 10.1146/annurev.genet.39.110304.095755. [DOI] [PubMed] [Google Scholar]
- 3.Hefferin ML, Tomkinson AE. Mechanism of DNA double-strand break repair by non-homologous end joining. DNA Repair. 2005;4:639–648. doi: 10.1016/j.dnarep.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 4.Baumann P, Cech TR. Protection of telomeres by the Ku protein in fission yeast. Mol. Biol. Cell. 2000;11:3265–3275. doi: 10.1091/mbc.11.10.3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boulton SJ, Jackson SP. Saccharomyces cerevisiae Ku70 potentiates illegitimate DNA double-strand break repair and serves as a barrier to error-prone DNA repair pathways. EMBO J. 1996;15:5093–5103. [PMC free article] [PubMed] [Google Scholar]
- 6.Manolis KG, Nimmo ER, Hartsuiker E, Carr AM, Jeggo PA, Allshire RC. Novel functional requirements for non-homologous DNA end joining in Schizosaccharomyces pombe. EMBO J. 2001;20:210–221. doi: 10.1093/emboj/20.1.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Teo SH, Jackson SP. Identification of Saccharomyces cerevisiae DNA ligase IV: involvement in DNA double-strand break repair. EMBO J. 1997;16:4788–4795. doi: 10.1093/emboj/16.15.4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feldmann E, Schmiemann V, Goedecke W, Reichenberger S, Pfeiffer P. DNA double-strand break repair in cell-free extracts from Ku80-deficient cells: implications for Ku serving as an alignment factor in non-homologous DNA end joining. Nucleic Acids Res. 2000;28:2585–2596. doi: 10.1093/nar/28.13.2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kabotyanski EB, Gomelsky L, Han JO, Stamato TD, Roth DB. Double-strand break repair in Ku86- and XRCC4-deficient cells. Nucleic Acids Res. 1998;26:5333–5342. doi: 10.1093/nar/26.23.5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kemp LM, Sedgwick SG, Jeggo PA. X-ray sensitive mutants of Chinese hamster ovary cells defective in double-strand break rejoining. Mutation Res. 1984;132:189–196. doi: 10.1016/0167-8817(84)90037-3. [DOI] [PubMed] [Google Scholar]
- 11.Smith J, Riballo E, Kysela B, Baldeyron C, Manolis K, Masson C, Lieber MR, Papadopoulo D, Jeggo P. Impact of DNA ligase IV on the fidelity of end joining in human cells. Nucleic Acids Res. 2003;31:2157–2167. doi: 10.1093/nar/gkg317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tzung TY, Runger TM. Reduced joining of DNA double strand breaks with an abnormal mutation spectrum in rodent mutants of DNA-PKcs and Ku80. Intl. J. Rad. Biol. 1998;73:469–474. doi: 10.1080/095530098142004. [DOI] [PubMed] [Google Scholar]
- 13.Ma JL, Kim EM, Haber JE, Lee SE. Yeast Mre11 and Rad1 proteins define a Ku-independent mechanism to repair double-strand breaks lacking overlapping end sequences. Mol. Cell. Biol. 2003;23:8820–8828. doi: 10.1128/MCB.23.23.8820-8828.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paull TT, Gellert M. A mechanistic basis for Mre11-directed DNA joining at microhomologies. Proc. Natl Acad. Sci. USA. 2000;97:6409–6414. doi: 10.1073/pnas.110144297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Audebert M, Salles B, Calsou P. Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III in an alternative route for DNA double-strand breaks rejoining. J. Biol. Chem. 2004;279:55117–55126. doi: 10.1074/jbc.M404524200. [DOI] [PubMed] [Google Scholar]
- 16.Wang H, Rosidi B, Perrault R, Wang M, Zhang L, Windhofer F, Iliakis G. DNA ligase III as a candidate component of backup pathways of nonhomologous end joining. Cancer Res. 2005;65:4020–4030. doi: 10.1158/0008-5472.CAN-04-3055. [DOI] [PubMed] [Google Scholar]
- 17.Wang M, Wu W, Wu W, Rosidi B, Zhang L, Wang H, Iliakis G. PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res. 2006;34:6170–6182. doi: 10.1093/nar/gkl840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.d’Adda di Fagagna F, Hande MP, Tong WM, Roth D, Lansdorp PM, Wang ZQ, Jackson SP. Effects of DNA nonhomologous end-joining factors on telomere length and chromosomal stability in mammalian cells. Curr. Biol. 2001;11:1192–1196. doi: 10.1016/s0960-9822(01)00328-1. [DOI] [PubMed] [Google Scholar]
- 19.Hsu HL, Gilley D, Galande SA, Hande MP, Allen B, Kim SH, Li GC, Campisi J, Kohwi-Shigematsu T, et al. Ku acts in a unique way at the mammalian telomere to prevent end joining. Genes Dev. 2000;14:2807–2812. doi: 10.1101/gad.844000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jaco I, Munoz P, Blasco MA. Role of human Ku86 in telomere length maintenance and telomere capping. Cancer Res. 2004;64:7271–7278. doi: 10.1158/0008-5472.CAN-04-1381. [DOI] [PubMed] [Google Scholar]
- 21.Myung K, Ghosh G, Fattah FJ, Li G, Kim H, Dutia A, Pak E, Smith S, Hendrickson EA. Regulation of telomere length and suppression of genomic instability in human somatic cells by Ku86. Mol. Cell Biol. 2004;24:5050–5059. doi: 10.1128/MCB.24.11.5050-5059.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Samper E, Goytisolo FA, Slijepcevic P, van Buul PP, Blasco MA. Mammalian Ku86 protein prevents telomeric fusions independently of the length of TTAGGG repeats and the G-strand overhang. EMBO Rep. 2000;1:244–252. doi: 10.1093/embo-reports/kvd051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karlseder J, Hoke K, Mirzoeva OK, Bakkenist C, Kastan MB, Petrini JH, de Lange T. The telomeric protein TRF2 binds the ATM kinase and can inhibit the ATM-dependent DNA damage response. PLoS Biol. 2004;2:E240. doi: 10.1371/journal.pbio.0020240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chan SW, Blackburn EH. Telomerase and ATM/Tel1p protect telomeres from nonhomologous end joining. Mol. Cell. 2003;11:1379–1387. doi: 10.1016/s1097-2765(03)00174-6. [DOI] [PubMed] [Google Scholar]
- 25.Celli GB, de Lange T. DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat. Cell Biol. 2005;7:712–718. doi: 10.1038/ncb1275. [DOI] [PubMed] [Google Scholar]
- 26.Ferreira MG, Cooper JP. The fission yeast Taz1 protein protects chromosomes from Ku-dependent end-to-end fusions. Mol. Cell. 2001;7:55–63. doi: 10.1016/s1097-2765(01)00154-x. [DOI] [PubMed] [Google Scholar]
- 27.Maser RS, Wong KK, Sahin E, Xia H, Naylor M, Hedberg HM, Artandi SE, DePinho RA. DNA-dependent protein kinase catalytic subunit is not required for dysfunctional telomere fusion and checkpoint response in the telomerase-deficient mouse. Mol. Cell Biol. 2007;27:2253–2265. doi: 10.1128/MCB.01354-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shakirov EV, Shippen DE. Length regulation and dynamics of individual telomere tracts in wild-type Arabidopsis. Plant Cell. 2004;16:1959–1967. doi: 10.1105/tpc.104.023093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fitzgerald MS, Riha K, Gao F, Ren S, McKnight TD, Shippen DE. Disruption of the telomerase catalytic subunit gene from Arabidopsis inactivates telomerase and leads to a slow loss of telomeric DNA. Proc. Natl Acad. Sci. USA. 1999;96:14813–14818. doi: 10.1073/pnas.96.26.14813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Riha K, McKnight TD, Griffing LR, Shippen DE. Living with genome instability: plant responses to telomere dysfunction. Science. 2001;291:1797–1800. doi: 10.1126/science.1057110. [DOI] [PubMed] [Google Scholar]
- 31.Heacock M, Spangler E, Riha K, Puizina J, Shippen DE. Molecular analysis of telomere fusions in Arabidopsis: multiple pathways for chromosome end-joining. EMBO J. 2004;23:2304–2313. doi: 10.1038/sj.emboj.7600236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Riha K, Shippen DE. Ku is required for telomeric C-rich strand maintenance but not for end-to-end chromosome fusions in Arabidopsis. Proc. Natl Acad. Sci. USA. 2003;100:611–615. doi: 10.1073/pnas.0236128100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Riha K, Watson JM, Parkey J, Shippen DE. Telomere length deregulation and enhanced sensitivity to genotoxic stress in Arabidopsis mutants deficient in Ku70. EMBO J. 2002;21:2819–2826. doi: 10.1093/emboj/21.11.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mathur J KC. Callus culture and regeneration. Methods Mol. Biol. 1998;82:31–34. doi: 10.1385/0-89603-391-0:31. [DOI] [PubMed] [Google Scholar]
- 35.Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 36.Gleave AP. A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol. Biol. 1992;20:1203–1207. doi: 10.1007/BF00028910. [DOI] [PubMed] [Google Scholar]
- 37.Friesner J, Britt AB. Ku80- and DNA ligase IV-deficient plants are sensitive to ionizing radiation and defective in T-DNA integration. Plant J. 2003;34:427–440. doi: 10.1046/j.1365-313x.2003.01738.x. [DOI] [PubMed] [Google Scholar]
- 38.Watson JM, Shippen DE. Telomere rapid deletion regulates telomere length in Arabidopsis thaliana. Mol. Cell Biol. 2007;27:1706–1715. doi: 10.1128/MCB.02059-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Churikov D, Wei C, Price CM. Vertebrate POT1 restricts G-overhang length and prevents activation of a telomeric DNA damage checkpoint but is dispensable for overhang protection. Mol. Cell Biol. 2006;26:6971–6982. doi: 10.1128/MCB.01011-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Attikum H, Bundock P, Overmeer RM, Lee LY, Gelvin SB, Hooykaas PJ. The Arabidopsis AtLIG4 gene is required for the repair of DNA damage, but not for the integration of Agrobacterium T-DNA. Nucleic Acids Res. 2003;31:4247–4255. doi: 10.1093/nar/gkg458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smogorzewska A, Karlseder J, Holtgreve-Grez H, Jauch A, de Lange T. DNA ligase IV-dependent NHEJ of deprotected mammalian telomeres in G1 and G2. Curr. Biol. 2002;12:1635–1644. doi: 10.1016/s0960-9822(02)01179-x. [DOI] [PubMed] [Google Scholar]
- 42.Jeggo PA. Identification of genes involved in repair of DNA double-strand breaks in mammalian cells. Radiation Res. 1998;150:S80–S91. [PubMed] [Google Scholar]
- 43.Tomita K, Matsuura A, Caspari T, Carr AM, Akamatsu Y, Iwasaki H, Mizuno K, Ohta K, Uritani M, et al. Competition between the Rad50 complex and the Ku heterodimer reveals a role for Exo1 in processing double-strand breaks but not telomeres. Mol. Cell Biol. 2003;23:5186–5197. doi: 10.1128/MCB.23.15.5186-5197.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goetz JD, Motycka TA, Han M, Jasin M, Tomkinson AE. Reduced repair of DNA double-strand breaks by homologous recombination in a DNA ligase I-deficient human cell line. DNA Repair. 2005;4:649–654. doi: 10.1016/j.dnarep.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 45.Bonatto D, Revers LF, Brendel M, Henriques JA. The eukaryotic Pso2/Snm1/Artemis proteins and their function as genomic and cellular caretakers. Brazilian J. Med. Biol. Res. 2005;38:321–334. doi: 10.1590/s0100-879x2005000300002. [DOI] [PubMed] [Google Scholar]
- 46.de Bruin D, Kantrow SM, Liberatore RA, Zakian VA. Telomere folding is required for the stable maintenance of telomere position effects in yeast. Mol. Cell Biol. 2000;20:7991–8000. doi: 10.1128/mcb.20.21.7991-8000.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de Bruin D, Zaman Z, Liberatore RA, Ptashne M. Telomere looping permits gene activation by a downstream UAS in yeast. Nature. 2001;409:109–113. doi: 10.1038/35051119. [DOI] [PubMed] [Google Scholar]
- 48.Grunstein M. Molecular model for telomeric heterochromatin in yeast. Curr. Opin. Cell. Biol. 1997;9:383–387. doi: 10.1016/s0955-0674(97)80011-7. [DOI] [PubMed] [Google Scholar]
- 49.Mengiste T, Revenkova E, Bechtold N, Paszkowski J. An SMC-like protein is required for efficient homologous recombination in Arabidopsis. EMBO J. 1999;18:4505–4512. doi: 10.1093/emboj/18.16.4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.