Abstract
Short interfering RNA (siRNA) duplexes are currently being evaluated as antisense agents for gene silencing. Chemical modification of siRNAs is widely expected to be required for therapeutic applications in order to improve delivery, biostability and pharmacokinetic properties. Beyond potential improvements in the efficacy of oligoribonucleotides, chemical modification may also provide insight into the mechanism of mRNA downregulation mediated by the RNA–protein effector complexes (RNA-induced silencing complex or RISC). We have studied the in vitro activity in HeLa cells of siRNA duplexes against firefly luciferase with substitutions in the guide strand of U for the apolar ribo-2,4-difluorotoluyl nucleotide (rF) [Xia, J. et al. (2006) ACS Chem. Biol., 1, 176–183] as well as of C for rF. Whereas an internal rF:A pair adjacent to the Ago2 (‘slicer’ enzyme) cleavage site did not affect silencing relative to the native siRNA duplex, the rF:G pair and other mismatches such as A:G or A:A were not tolerated. The crystal structure at atomic resolution determined for an RNA dodecamer duplex with rF opposite G manifests only minor deviations between the geometries of rF:G and the native U:G wobble pair. This is in contrast to the previously found, significant deviations between the geometries of rF:A and U:A pairs. Comparison between the structures of the RNA duplex containing rF:G and a new structure of an RNA with A:G mismatches with the structures of standard Watson–Crick pairs in canonical duplex RNA leads to the conclusion that local widening of the duplex formed by the siRNA guide strand and the targeted region of mRNA is the most likely reason for the intolerance of human Ago2 (hAgo2), the RISC endonuclease, toward internal mismatch pairs involving native or chemically modified RNA. Contrary to the influence of shape, the thermodynamic stabilities of siRNA duplexes with single rF:A, A:A, G:A or C:A (instead of U:A) or rF:G pairs (instead of C:G) show no obvious correlation with their activities. However, incorporation of three rF:A pairs into an siRNA duplex leads to loss of activity. Our structural and stability data also shed light on the role of organic fluorine as a hydrogen bond acceptor. Accordingly, UV melting (TM) data, osmotic stress measurements, X-ray crystallography at atomic resolution and the results of semi-empirical calculations are all consistent with the existence of weak hydrogen bonds between fluorine and the H-N1(G) amino group in rF:G pairs of the investigated RNA dodecamers.
INTRODUCTION
Messenger RNAs (mRNAs) represent a necessary step in gene expression, being the intermediary between gene and protein. Short interfering RNAs (siRNAs) are duplexes of RNA, 21–23 nt long, and consist of the sense and the guide strand. The latter becomes incorporated into a complex of proteins and serves to identify a complementary sequence in an mRNA. This either causes the mRNA to be cleaved by the protein complex or prevents it from being translated into protein (1–8), thus effectively silencing the encoding gene. Therefore, RNA interference (RNAi) is a powerful biological process for specific silencing of gene expression in diversified eukaryotic cells and has tremendous potential for functional genomics, drug discovery through in vivo target validation, and development of novel gene-specific medicine. It is hoped that this approach might be used to shut down disease-related genes in humans.
The RNAi process within cells comprises at least four sequential steps (9–15): (i) ATP-dependent processing of double-stranded RNA into siRNAs by ‘Dicer’, an RNase III-family enzyme; (ii) incorporation of these naturally derived siRNAs, or synthetic siRNAs introduced into the cell, into an inactive ribonucleoprotein complex; (iii) ATP-dependent unwinding of the siRNA duplex to generate an active RNA-induced silencing complex (RISC) whereby ATP is used to maintain 5′-phosphates on siRNAs and (iv) cleavage of the RNA target in an ATP-independent reaction. RISC is a sequence-specific endonuclease complex that contains Argonaute (Ago) proteins and the single-stranded guide siRNA strand. RISC mediates cleavage of target RNAs with multiple turnovers in the absence of additional ATP. The high specificity of the process is based on Watson–Crick pairing between the siRNA antisense (guide) oligonucleotide and the target mRNA. Within the RISC complex, the Ago2 enzyme is exclusively responsible for cleavage of the mRNA (16,17). Structural data indicate that the PIWI domain of Ago2 that adopts an RNase H-like fold is responsible for the ‘Slicer’ activity (18).
The cleavage position on the mRNA in a 21-mer siRNA duplex with mRNA is located between nucleotides 10 and 11 upstream from the target nucleotide paired to the 5′-terminus of the antisense strand (12). The sites of active duplexes are not predictable, but numerous siRNAs targeted against a particular message have been studied in attempts to correlate duplex features with function. An evaluation of the activity of 25 duplexes targeting the open reading frame and 3′-untranslated region of lamin A/C found no bias based on position within the message (19). Variations in siRNA efficiency as a function of small positional shifts on the target (19,20) could be due to competition for binding with protein factors, different thermodynamic stabilities, or sequence dependence of the RISC complex. In addition, the structure and accessibility of the target message will also play an important role (21) and prediction algorithms may offer some guidance for siRNA sequence selection (22). The most effective siRNAs have a tendency to pair more weakly at the 5′-antisense side of the duplex relative to the 3′-antisense side (22,23). Several criteria including base composition, strength of base pairing at particular positions in the duplex, overall thermodynamic stability and base identities were used to select siRNA duplexes against five genes (24). Duplexes that were selected based on these criteria resulted in a mean silencing of 76% compared to 39% mean silencing for those that were randomly selected. Introduction of mismatches in the central region of the antisense strand was found to abolish silencing activity (12,25), but the sense strand can tolerate limited mismatches (25,26). Interestingly, when mismatches were placed in the terminal four-base region between the 5′-end of the antisense strand and the 3′-end of the sense strand improvements in activity were observed (27). Chemically modified siRNAs linked by a hairpin loop can still trigger target RNA cleavage through the RNAi pathway (19,28–30). However, it appears that recruitment by RISC requires siRNA duplexes because most antisense-only, single-stranded siRNAs show much less activity than the corresponding duplexes (10). Approaches to the successful identification of effective and specific siRNAs have recently been reviewed (25).
Chemical modification of siRNA duplexes on one or both strands will likely be required to improve their efficacy for in vivo applications despite the fact that short double-stranded RNAs show relatively high stability and activity in cell culture (31–38). Chemical modification is desirable to enhance: (i) the siRNA-binding affinity by, for example, conformationally preorganizing the guide strand for the mRNA; (ii) the nuclease stability and therefore the half-life of the siRNA duplexes in circulation in vivo and (iii) the biodistribution and pharmacokinetic properties of siRNAs and to allow targeting of siRNA to certain cell types. Incorporation of phosphorothioate (PS) linkages, 2′-OMe- or 2′-fluoro-nucleotides into siRNAs was found to affect their activities dependent on the position and extent of the modifications (35–38). For example, siRNA duplexes with the sense or antisense strand fully 2′-OMe-modified showed a significantly diminished activity in HeLa cells despite efficient silencing by the corresponding native RNAs (39). And siRNAs with both strands fully 2′-OMe-modified were inactive. On the other hand siRNAs that featured fully PS- and 2′-OMe-modified sense strands displayed similar efficacy and potency as the unmodified parent duplexes in some cases (40). The limited activity seen in some cases for duplexes with completely modified antisense strands may therefore relate to altered unwinding of the modified duplex or loading of the unmodified antisense-strand into RISC rather than inaccessibility of the target.
We recently evaluated the in vitro activity of siRNAs containing apolar ribo-2,4-difluorotoluyl nucleotides (rF) in place of U (Figure 1A and B) (41). Modification of the guide strand at the 5′-end with rF was found not to affect silencing compared with the unmodified control. Moreover, siRNAs with an internal rF modification, despite a slightly reduced affinity for their target effectively silenced gene expression and they also exhibited improved nuclease resistance in serum. A RACE analysis demonstrated that the siRNA-mediated cleavage occurred at the predicted site both for the unmodified and the rF-containing duplex. The crystal structure of an RNA duplex with rF:A pairs revealed local conformational variations relative to canonical RNA (41). rF and A paired more loosely compared with U and A and there was an absence of ordered water molecules around the difluorotoluyl moiety in both grooves. Whereas substitution of rF for U essentially had no effect on silencing, base mismatches at the corresponding positions led to a loss of (G:A, A:A) or reduced (C:A) activity (41).
Figure 1.
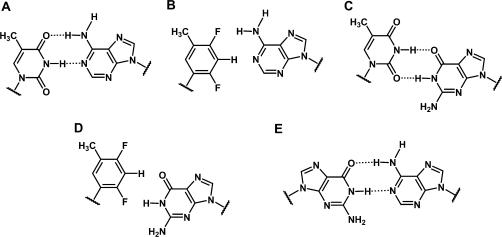
Chemical structures of base pairs. (A) T:A; (B) F:A; (C) U:G; (D) F:G; (E) G:A. Hydrogen bonds are indicated by dashed lines.
Here, we report in vitro activity data of siRNAs with U:G or rF:G pairs (Figure 1C and D) and crystal structures near 1 Å-resolution of RNA dodecamer duplexes with either rF:G (FG 12-mer) or A:G pairs (GA 12-mer) (Figure 1E). To further investigate the relative stability of T:A, rF:A, T:G and rF:G pairs we have carried out semi-empirical calculations at the MP2/6-31G*and MP2/6-31+G** levels. The combined data allow insights into substrate specificity by hAgo2 and indicate that RNA duplex shape at or near the cleavage site is a more important determinant of RISC-mediated activity of siRNAs than thermodynamic stability.
MATERIALS AND METHODS
Oligoribonucleotide synthesis and purification
The rF phosphoramidite building block for incorporation into RNAs was prepared as described in ref. (41). All siRNA molecules and the rF-containing dodecamers for stability and crystallographic studies were synthesized on a 394 ABI synthesizer using previously described protocols (41). The oligonucleotides were cleaved from the CPG, with simultaneous deprotection of base and phosphate groups, with 1.0 ml of a mixture of ethanolic ammonia (ammonia–ethanol, 3:1) for 16 h at 55°C. The solution was decanted, lyophilized and re-suspended in 0.2 ml of triethylamine trihydrofluoride (TEA.3HF, Aldrich) and was incubated at 65°C for 90 min to remove the t-butyldimethylsilyl (TBDMS) groups. The completely deprotected oligonucleotides were then precipitated from anhydrous methanol (MeOH, 0.4 ml). The oligonucleotides were purified by PAGE (41) and the desired bands were excised and shaken overnight in 5 ml of 100 mM sodium acetate. The extracted oligonucleotides were desalted using C-18 Sep-Pak cartridges (Waters). The oligonucleotides were characterized by electrospray mass spectroscopy (ES/MS) and analytical anion-exchange HPLC and/or capillary gel electrophoresis (CGE).
The GA dodecamer r(CGCGAAUUAGCG) was synthesized on a Pharmacia Gene Assembler Plus instrument using 2′-O-TOM nucleoside phosphoramidites (42) and previously described protocols (43). Deprotection and cleavage from the solid support was achieved in a mixture of MeNH2 in EtOH (8 M, 0.6 ml) and MeNH2 in H2O (40%, 0.6 ml) for 5 h. After the solution was completely evaporated, tetrabutylammonium fluoride trihydrate (TBAF.3H2O) in THF (1 M, 0.95 ml) was added. The reaction mixture was slowly shaken for 14 h at room temperature to remove the 2′-O-silyl ethers. The reaction was quenched by the addition of triethylammonium acetate (TEAA) (1 M, pH 7.4, 0.95 ml). The volume of the solution was reduced to 1 ml and the solution was loaded on a Amersham HiPrep 26/10 Desalting column (2.6 × 10 cm; Sephadex G25). The crude RNA was eluted with H2O and dried. Analysis of crude RNA products after deprotection was performed by anion-exchange chromatography on a Dionex DNAPac100 column (4 × 250 mm) at 80°C. Flow rate: 1 ml/min; eluant A: 25 mM Tris–HCl (pH 8.0), 6 M urea; eluant B: 25 mM Tris–HCl (pH 8.0), 0.5 M NaClO4, 6 M urea; gradient: 0–60% B in A within 45 min; UV-detection at 265 nm. The crude RNA (trityl-off) was purified on a semi-preparative Dionex DNAPac100 column (9 × 250 mm). Flow rate: 2 ml/min; gradient: 5–10% B in A within 20 min. Fractions containing RNA were loaded on a C18 SepPak cartridge (Waters/Millipore), washed with 0.1–0.2 M (Et3NH)HCO3, H2O, and eluted with H2O/CH3CN (6/4). RNA fractions were lyophilized.
In vitro analysis of luciferase expression
HeLa cells that stably express both firefly and Renilla luciferase (HeLa Dual-luc cells) were grown as previously described (41). In vitro activity of siRNAs was determined using a high-throughput 96-well plate format assay for luciferase activity. HeLa Dual-luc cells were transfected with firefly luciferase targeting siRNAs at concentrations ranging from 0.04 to 30 nM using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. After 24 h, cells were analyzed for both firefly and Renilla luciferase expression using a plate luminometer (VICTOR2, PerkinElmer) and the Dual-Glo Luciferase Assay kit (Promega). Firefly/Renilla luciferase expression ratios were used to determine the percentage of gene silencing relative to untreated controls.
Thermal denaturation (TM) and osmotic stress studies
Melting of each oligonucleotide (4 μM) was done in 10 mM sodium cacodylate, 0.1 mM EDTA and 300 mM NaCl in presence of 0, 5, 10, 15 and 20% weight/volume of each of the three organic co-solutes in Table 1. Oligonucleotide concentrations were calculated from the nearest-neighbor approximation (44) using ε = 1000 M−1 cm−1 (λ = 260 nm) as the extinction coefficient for rF nucleoside (45). Absorbance versus temperature profiles were measured at 280 nm on a Varian Bio 100 spectrometer equipped with a six-position Peltier temperature controller. The temperature was increased at 0.5°C per minute. Five samples were measured concurrently in the double-beam mode. The melting temperatures and thermodynamic parameters were obtained using Varian Cary software (Version 02.00). The experimental absorbance versus temperature curves were converted into a fraction of strands remaining hybridized (α) versus temperature curves by fitting the melting profile to a two-state transition model, with linearly sloping lower and upper base lines. The melting temperatures (TM) were obtained directly from the temperature at α = 0.5. The final TM was an approximation of usually five to eight measurements.
Table 1.
Thermodynamic stability (TM) and osmotic stress data for self-complementary RNA 12-mers with U:G, rF:A, rF:G and A:G pairs
| Sequence | Name | TaM (°C) | ▵nw Ethylene glycol | ▵nw Glycerol | ▵nw Acetamide |
|---|---|---|---|---|---|
| CGCUAAUUGGCG | UG | 57.7 ± 0.2 | 18.5 ± 3.0 | 20.3 ± 3.4 | 33.1 ± 3.0 |
| CGCFAAUUGGCG | FG | 54.9 ± 0.2 | 14.6 ± 2.1 | 26.0 ± 3.3 | 29.2 ± 2.4 |
| CGCGAAUUFGCG | GF | 53.9 ± 0.3 | 13.5 ± 2.4 | 24.7 ± 3.7 | 29.6 ± 3.1 |
| CGCFAAUUAGCGb | FA | 53.2 ± 0.3 | 13.2 ± 3.1 | 12.7 ± 3.3 | 23.6 ± 2.9 |
| CGCGAAUUAGCG | GA | 58.2 ± 0.2 | 21.2 ± 2.6 | 23.8 ± 3.1 | Not measured |
| CGCAAAUUUGCGc | AU | 58.7 ± 0.6 | 22.8d | 18d | 43.2d |
The thermodynamic parameters were determined from the width at the half-height of the differentiated melting curve. The fraction of strands remaining hybridized (α) versus temperature curves were converted into differentiated melting curves [δα/δ(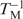 ) versus TM] using Varian Cary software (Version 02.00). The width of the differentiated melting curve at the half-height is inversely proportional to the van't Hoff transition enthalpy; for a bimolecular transition ΔH = 10.14/(
) versus TM] using Varian Cary software (Version 02.00). The width of the differentiated melting curve at the half-height is inversely proportional to the van't Hoff transition enthalpy; for a bimolecular transition ΔH = 10.14/( ) where T1 is the lower temperature and T2 is the upper temperature (both in K) at one-half of [δα/δ(
) where T1 is the lower temperature and T2 is the upper temperature (both in K) at one-half of [δα/δ(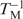 )] (46).
)] (46).
The changes in the number of water molecules associated with the melting process ▵nw were determined as described by Spink and Chaires (47) ▵nw = (−▵H/R)[d(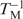 )/d(lnaw)], where −▵H is the enthalpy determined from the width at the half-height of differentiated melting curve in pure buffer and R is the universal gas constant (1.986 calmol−1 K−1). The experimentally determined values of water activity (lnaw) at given co-solute concentrations were provided by Professors Spink and Chaires. The slope of the plot of reciprocal temperature (in K) of melting versus the logarithm of water activity (lnaw) at different concentrations (0, 5, 10, 15 and 20%) of small co-solutes gave the value of d(
)/d(lnaw)], where −▵H is the enthalpy determined from the width at the half-height of differentiated melting curve in pure buffer and R is the universal gas constant (1.986 calmol−1 K−1). The experimentally determined values of water activity (lnaw) at given co-solute concentrations were provided by Professors Spink and Chaires. The slope of the plot of reciprocal temperature (in K) of melting versus the logarithm of water activity (lnaw) at different concentrations (0, 5, 10, 15 and 20%) of small co-solutes gave the value of d(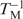 )/d(lnaw). The final ▵nw were obtained by linear fitting using KaleidaGraph software (Version 3.51) with a confidence level usually better than 98%. The experimental uncertainties were obtained as previously reported (48).
)/d(lnaw). The final ▵nw were obtained by linear fitting using KaleidaGraph software (Version 3.51) with a confidence level usually better than 98%. The experimental uncertainties were obtained as previously reported (48).
Crystallization of RNA dodecamers containing rF:G and A:G pairs and X-ray diffraction data collection
Crystals of the FG dodecamer r(CGCFAAUUGGCG) were grown at 18°C by the sitting-drop vapor diffusion method, using the Grid Screen Ammonium Sulfate (Hampton Research Inc., Aliso Viejo, CA, USA). Droplets containing 0.5 mM oligonucleotide, 0.8 M ammonium sulfate and 0.05 M MES, pH 6.0, were equilibrated against a reservoir of 1.6 M ammonium sulfate, 0.1 M MES, pH 6.0. Crystals appeared after 3 days. Crystals of the AG dodecamer r(CGCGAAUUAGCG) were grown at 18°C by the hanging-drop vapor diffusion method, using the Nucleic Acid Miniscreen (Hampton) (49). Both conditions 14 and 16 resulted in crystals. Crystals for diffraction data collection were grown from droplets containing 0.5 mM oligonucleotide, 5% 2-methyl-2,4-pentanediol (MPD), 20 mM sodium cacodylate, pH 7.0, 6 mM spermine-4HCl, 40 mM sodium chloride and 10 mM magnesium chloride that were equilibrated against a reservoir of 35% MPD. Crystals appeared after 2 days. In both cases, crystals were mounted in nylon loops and frozen in liquid nitrogen. Diffraction data were collected at 120 K on the insertion device beamlines of the Southeast Regional Collaborative Access Team (SER-CAT, 22-ID, FG dodecamer) and the DuPont–Northwestern–Dow Collaborative Access Team (DND-CAT, 5-ID, GA dodecamer) at the Advanced Photon Source (APS), Argonne, IL. The wavelengths for the data collections varied between ca. 0.9 and 1.0 Å. In both cases high- and low-resolution data sets were collected separately. Diffraction data were processed with the programs HKL2000 (50) (FG) and XDS (51) (GA).
Structure determination and refinement
Both structures were determined by the molecular replacement method using the program EPMR (52). For the FG RNA structure, a canonical A-RNA dodecamer constructed with the program TURBO-FRODO (53) was used as the search model. A previously published structure at low resolution (54) served as the search model for structure determination of the GA RNA duplex. In both cases, the structures were initially refined with CNS (55) and at a later stage with the program SHELX-97 (56). In the subsequent refinement cycles, solvent water molecules were added, and anisotropic temperature factors refinement was carried out. The program TURBO-FRODO was used to display electron density maps for manual rebuilding of the models. To calculate the R-free, 5% randomly chosen reflections were set aside in both cases.
Computational methods
Ab initio calculations were carried out using the GAUSSIAN 03 code (57) on a 64-bit Linux machine. Optimizations were initiated at the HF-3-21G level, followed by calculations at the MP2/6-31G* and finally at the MP2/6-31+G** levels. No constraints, i.e. to maintain planarity of the base pairs were used. Interaction energies were computed as the difference in energy between the optimized base pair on one hand, and the sum of energies of optimized components on the other. To calculate the point energies of base pairs based on the crystallographic conformations, hydrogen atoms were added by the program. All quantities were corrected for basis set superposition error by the standard counterpoise procedure (58).
Coordinates
Final coordinates and structure factors for the FG and GA RNAs have been deposited in the Protein Data Bank (http://www.rcsb.org.) The PDB ID codes are 2Q1O (FG duplex) and 2Q1R (GA duplex).
RESULTS
Gene silencing activity of siRNAs with U or C replaced by rF
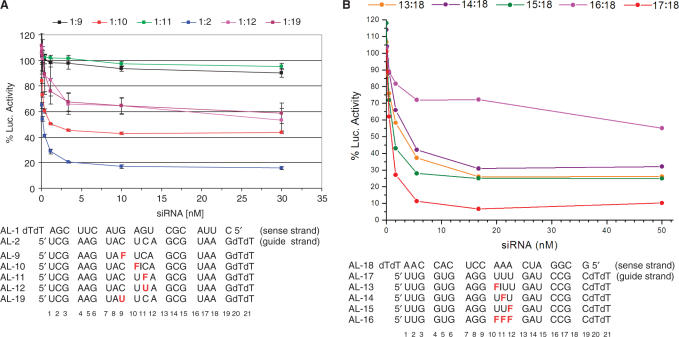
To examine the effects on gene silencing of rF incorporation into siRNAs, we employed an in vitro firefly luciferase assay in HeLa cells. Two regions of the luciferase gene were targeted separately with RNA duplexes whose guide strands contained single or multiple rF residues (Figure 2) (41). The silencing activities of modified duplexes applied in concentrations ranging between 0.04 and 30 nM were measured in the form of a decreased luciferase luminescence and compared with the signals resulting from treatment with native control siRNA duplexes. A single uridine positioned three or more nucleotides upstream or downstream from the cleavage site in the guide strand could be replaced by rF without a significant change in the 50% inhibitory concentration (IC50) (41). Moreover, replacement of the U adjacent to the cleavage site by rF only increased the IC50 from 0.21 to 1.2 nM (Figure 2A). However, substitution of either C9 or C11 for U or rF, resulting in the formation of U:G and rF:G pairs, respectively, significantly diminished or even abolished the silencing activity altogether (IC50 > 30 nM; Figure 2A). Similarly, replacement of U10 by A or G resulted in a loss of activity, whereas a U10→C substitution (leading to a C:A mismatch) yielded an IC50 of 0.92 nM, comparable to the activity of an siRNA duplex with the U10→rF substitution (41). Therefore, it appears that a single purine–purine mismatch pair or U:G/rF:G wobble pairs (assuming the rF:G pair adopts a wobble geometry) at or adjacent to the cleavage site are not tolerated by the hAgo2 endonuclease. The C:A mismatch and rF:A constitute noteworthy exceptions. A RACE analysis of the cleavage products had established that the position of the cleavage site remained unchanged as a result of using siRNA duplexes with single rF:A pairs compared with native siRNAs (41).
Figure 2.
Concentration-dependent in vitro silencing activity of siRNA duplexes directed against firefly luciferase relative to Renilla luciferase. (A) Incorporation of a single rF residue opposite A in the sense [data not shown, see ref. (41)] and guide strands (1:10) is tolerated albeit with a decrease in activity for the latter (reference duplex 1:2). On the other hand, incorporation of rF or U opposite G is not or significantly less tolerated (1:9, 1:11 and 1:12, 1:19, respectively). (B) Contrary to the presence of a single rF residue in the guide strand opposite A (13:18, 14:18, 15:18; reference duplex 17:18), multiple rF:A pairs in an siRNA duplex prevent silencing (16:18). In the AL-10, AL-11, AL-13 and AL-14 oligonucleotides, the rF substitution is adjacent to the cleavage site (marked by a vertical line).
Whereas a single rF residue in place of U was tolerated in the guide strand, consecutive replacement of three Us also led to a loss of activity (Figure 2B). The thermodynamic stabilities (UV melting TM's) of siRNA 21-mer duplexes with single rF:A, A:G, A:A or C:A pairs are comparable (between 65 and 67°C relative to the native construct = 73°C) (41). Therefore, the observed difference in silencing activity between siRNA duplexes containing rF:A or any of the above mismatch pairs is unlikely to be a consequence of reduced stability. Conversely, three consecutive rF:A pairs may lead to local melting of the RNA duplex and considerably distort or destroy the secondary and tertiary structure of the hAgo2 substrate.
Thermodynamic stability of and osmotic stress measurements for oligoribonucleotides with U→rF or C→rF substitutions
To assess the effect of rF incorporation opposite A or G on the thermodynamic stability of an RNA dodecamer duplex we carried out UV melting experiments. Initial runs at a wavelength of 260 nm showed nonlinear upper or lower base lines, a problem that was eliminated by measuring optical densities as a function of temperature at 280 nm. The resulting TM's are listed in Table 1. For the comparison here between the six sequences we have listed TM differences per modified base pair (note that there are two substitutions per self-complementary dodecamer duplex). Remarkably, substituting U for rF opposite G resulted in a TM reduction of just 1.4°C per modification (UG versus FG duplex). In the case of the dodecamer with G and rF switched (GF) the reduction in TM relative to the UG duplex amounts to 1.9°C per modification. Switching a U:G (UG) to an rF:A pair (FA) lowers the TM by 2.3°C. Therefore, the rF:G is more stable than the rF:A pair (▵TM = 0.9°C; FG versus FA duplex). The difference in TM between the FA and A:U pairs (AU) is 2.8°C. For completeness, we also measured the stability of the GA duplex with two G:A mismatch pairs. The GA duplex was slightly less stable than the AU duplex (▵TM = −0.3°C; GA versus AU duplex) but slightly more stable than the UG duplex (▵TM = 0.3°C; GA versus UG duplex). Overall the observed order of stability of the pairs at pH 6.5 is: A:U > G:A > U:G > rF:G > G:rF > rF:A.
In order to gain insight into changes in RNA hydration as a result of rF incorporation, we employed the osmotic stress method (47,59). The reductions in the RNA melting temperatures upon addition of ethylene glycol, glycerol or acetamide were measured at different concentrations of these small co-solutes (48,60). The changes in the number of water molecules ▵nw associated with the melting process were determined as described by Spink and Chaires (47): ▵nw = (−▵H/R)[d(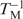 )/d(lnaw)], where ▵H is the measured enthalpy, R is the universal gas constant (1.986 calmol−1 K−1) and lnaw are experimentally determined values of water activity at given co-solute concentrations. This approach to establish the number of water molecules bound to a nucleic acid duplex is based on the assumption that the co-solutes (ethylene glycol, glycerol, acetamide) only change the water activity and do not directly interact with the RNA.
)/d(lnaw)], where ▵H is the measured enthalpy, R is the universal gas constant (1.986 calmol−1 K−1) and lnaw are experimentally determined values of water activity at given co-solute concentrations. This approach to establish the number of water molecules bound to a nucleic acid duplex is based on the assumption that the co-solutes (ethylene glycol, glycerol, acetamide) only change the water activity and do not directly interact with the RNA.
Rather than interpreting the absolute values obtained for ▵nw we believe it may be more instructive to consider the relative differences in amounts of water released when comparing the duplexes (Table 1). Interestingly, in the ethylene glycol and acetamide series the general trend regarding hydration followed the above order of thermodynamic stabilities (TM): A:U > G:A > U:G > rF:G > G:rF > rF:A. Thus, introduction of the G:A and U:G mismatches gradually decreased the hydration. Introduction of rF as a base-pairing partner decreased ▵nw even more, although the effect may be considered a relatively minor one, especially if one considers the highly hydrophobic nature of the rF modification. In all three series, hydration of the duplexes with either rF:G or G:rF pairs was more favorable than the hydration of the duplex with rF:A pairs, which is in agreement with the observations made in crystal structures (vide infra). Overall, the trends were less clear in the glycerol series. As previously observed (48,60), the ▵nw values were somewhat higher in the acetamide than in the ethylene glycol or glycerol series. A possible explanation for such discrepancies might be the fact that the organic co-solutes display some interaction with the RNA (61,62). Record and coworkers (62) showed that most small organic co-solutes (including glycerol) were not completely excluded from the surface of bovine serum albumin. When used in osmotic stress experiments, such co-solutes are expected to underestimate the magnitude in change of hydration (62). Whereas the inertness of the co-solutes and reliability of the absolute numbers of ▵nw is therefore still under debate (61,62), we (48,60) and others (47) have used the relative trends to gain insight into the changes in hydration in native and modified nucleic acids.
X-ray crystallography of RNA dodecamers with rF:G and A:G mismatches
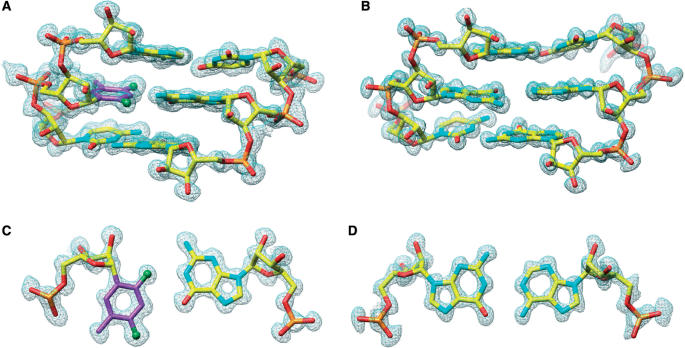
In order to gain a better understanding of the in vitro siRNA activity and thermodynamic stability/hydration data of RNAs containing rF residues and to compare the geometry of the rF:G pair with that of the previously visualized rF:A pair (41), we determined the crystal structure at 1.10 Å resolution of the RNA dodecamer duplex [r(CGCFAAUUGGCG)]2 with rF:G pairs (FG duplex, Table 1). The structure of the RNA dodecamer duplex [r(CGCGAAUUAGCG)]2 with A:G mismatch pairs (GA duplex, Table 1) at 1.12 Å was determined to potentially provide a structural rationale for the inactivity of siRNA duplexes with an incorporated mismatch pair at the cleavage site in in vitro assays. A summary of crystal data and refinement parameters for both structures is provided in Table 2. Sum (2Fo–Fc) electron density maps based on the final models of the FG and GA duplexes around selected regions are depicted in Figure 3.
Table 2.
Crystal data, data collection parameters and structure refinement statistics
| Structure | 5′-CGCFAAUUGGCG-3′ (FG) | 5′-CGCGAAUUAGCG-3′ (GA) |
|---|---|---|
| Crystal data | ||
| Space group | P1 | C2 |
| Number of strands per asym. unit | 4 | 1 |
| Cell dimensions | ||
| a, b, c (Å) | 22.90, 32.06, 42.93 | 41.34, 34.89, 32.12 |
| α, β, γ (°) | 96.2, 83.9, 93.9 | 90.0, 129.3, 90.0 |
| Data collection | ||
| Number of reflections | 45 752 | 13 514 |
| Resolution (last shell) | 1.10 (1.14–1.10) | 1.12 (1.16–1.12) |
| Completeness (%, last shell) | 93.9 (90.7) | 96.4 (89.3) |
| Rmerge (last shell) | 0.059 (0.239) | 0.035 (0.277) |
| Refinement | ||
| Rwork/Rfree | 0.112/0.142 | 0.147/0.187 |
| Number of atoms | ||
| RNA | 1016 | 255 |
| Water | 324 | 76 |
| Ions | – | 2 Mg2+ |
| B-factors | ||
| Nucleic acid (Å2) | 9.9 | 13.4 |
| Water (Å2) | 29.0 | 29.7 |
| r.m.s.d. | ||
| Bond lengths (Å) | 0.014 | 0.014 |
| Bond angles (1···3; Å) | 0.029 | 0.031 |
Figure 3.
Quality of the final Fourier (2Fo–Fc) sum electron density around the RNA duplexes in the region of the chemically modified or mismatched base pairs. (A) Portion of the FG duplex viewed into the minor groove; (B) portion of the GA duplex viewed into the minor groove; (C) rF104:G209 in the FG duplex and (D) G104:A209 in the GA duplex. Atoms are colored yellow, red, cyan and orange for carbon, oxygen, nitrogen and phosphorus, respectively. Carbon atoms of the difluorotoluyl moiety are highlighted in magenta and fluorine atoms are shown as green spheres.
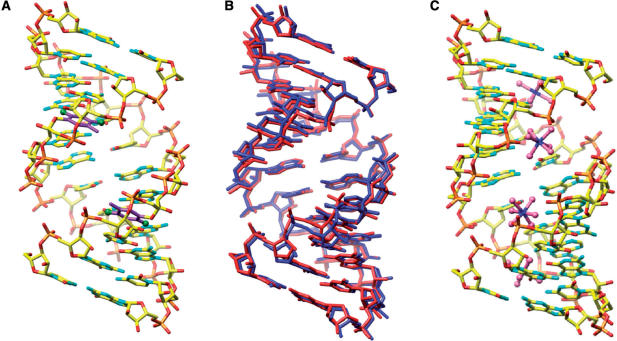
The geometries of the FG and GA duplexes (all riboses adopt a C3′-endo pucker) deviate from a more or less canonical A-form only in the vicinity of the rF:G and A:G pairs (Figure 4). rF:G pairs exhibit the shear typical for the U:G wobble pair whereby the guanine and difluorotoluene moieties are pushed into the minor and major grooves, respectively (Figures 1C and 3C). In the GA duplex, A and G are both in the standard ‘anti’ conformation and pair in a pseudo Watson–Crick fashion (Figures 1E and 3D). As a result, the minor groove of the GA duplex is widened by ∼1 Å with a concomitant narrowing of the major groove. The FG structure features two independent duplexes per crystallographic asymmetric unit, leading to four observations of the rF:G pair. Both duplexes have very similar geometries as revealed by a superimposition (Figure 4B). The structure of the GA duplex was previously reported at a lower resolution by others (1.9 Å) (54). However, in that structure with space group P21 the asymmetric unit consisted of a single duplex in a general position (the presence of a strong non-crystallographic 2-fold symmetry was noted by the authors at the time). Conversely, the space group of the GA RNA crystals analyzed here is C2, but the unit cell constants are virtually identical to those reported earlier. The conversion from pseudo-2-fold to crystallographic 2-fold symmetry is most likely a consequence of different crystallization conditions (see Materials and Methods section). In the C2 structure, the duplex is located on a crystallographic dyad and the asymmetric unit consists of a single dodecamer strand (Table 2). Summaries of the helical parameters calculated with the program CURVES (63) for the two FG duplexes and the GA duplex are provided in the Supplementary Data. Residues in the GA duplex are numbered C101 to G112 and residues in the four independent strands of the FG duplexes are numbered C101 to G112 (strand 1), C201 to G212 (strand 2), C301 to G312 (strand 3) and C401 to G412 (strand 4) (Figures 3 and 5).
Figure 4.
Overall conformation of RNA dodecamer duplexes with rF:G pairs. (A) One of the two RNA duplexes (duplex 1) per crystallographic asymmetric unit viewed into the major groove. Carbon atoms of the F moiety are highlighted in magenta and fluorine atoms are shown as green spheres. (B) Superimposition of the two duplexes; duplex 1 is red and duplex 2 is blue. (C) Overall conformation of the RNA dodecamer duplex with A:G pairs; the view is into the major groove that contains four Mg2+ hexahydrate complexes.
Figure 5.
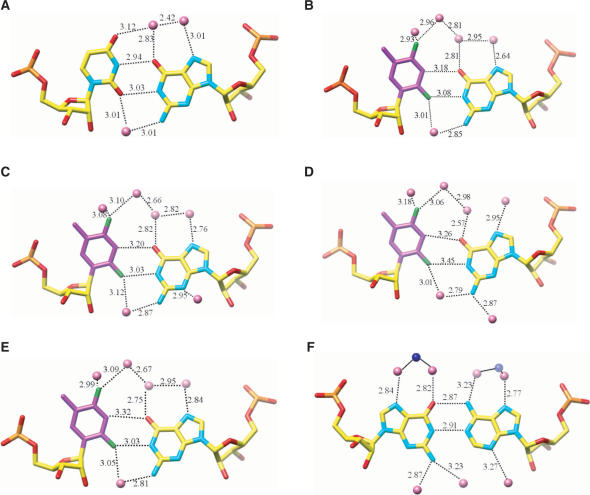
Geometry and hydration of rF:G, U:G and G:A base pairs. (A) U:G pair in structure NDB AR0009 (66); (B) rF104:G209; (C) rF204:G109; (D) rF304:G409; (E) rF404:G309; (F) G104:A209# (# designates a symmetry-related residue). Hydrogen bonds in the U:G and G:A base pairs and the corresponding interactions in the rF:G pairs are shown as dashed lines with distances in Å. Water molecules and Mg2+ ions are pink and blue spheres, respectively.
The four rF:G pairs exhibit similar geometries (Figure 5); the (rF)C3-H … O6(G) hydrogen bond distance varies between 3.18 and 3.31 Å. In three of the pairs the (rF)F2 … N1(G) distance is below 3.1 Å and in the rF304:G409 pair this distance is somewhat longer (3.45 Å, Figure 5D). Therefore, the fluorine–amine interaction here is considerably shorter than the sum of the van der Waals radii. If we assume the radii of fluorine and hydrogen to be 1.35 Å and 1.2 Å, respectively (64), and a N-H distance of 1 Å, the F … N distances in three of the four rF:G pairs are ca 0.5 Å below the sum (3.55 Å). The wisdom of assigning a larger radius to fluorine than to hydrogen has been questioned recently (65). Still, even we assume the same radius for fluorine and hydrogen (i.e. 1.2 Å), the observed distances between F2 and N1 in the rF:G pairs are indicative of an attractive interaction. The shortest distance (3.03 Å) between F2(rF) and N1(G) seen here is actually more than 0.8 Å shorter than the average distance between F4(rF) and N6(A) in rF:A pairs (41)! This difference is noteworthy because the distances of the (rF)C3-H … O6(G)/N1(A) hydrogen bonds in the rF:G and rF:A pairs are very similar. The close resemblance of the rF:G and U:G pairs (Figure 5) can itself be taken as evidence for an attractive interaction between fluorine and N1-H(G). If the (rF)C3-H … O6(G) hydrogen bond provided the only or the main contribution to the stability of the rF:G pair, the shear seen between F and G would seem quite unnecessary. Likewise, shearing does not benefit stacking interactions; if the geometry of rF:G and rF:A pairs were chiefly determined by stacking it would be difficult to explain their different geometries.
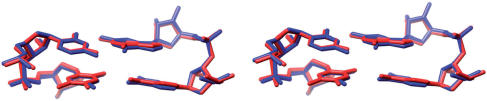
The similarity between rF:G and U:G is not limited to pairing geometry but extends to nearest neighbor interactions and hydration. Thus, superimposition of (CpU):(GpG) and (CprF):(GpG) base-pair steps reveals virtually identical conformations (Figure 6). The four rF:G pairs in the FG structure display a conserved hydration pattern (Figure 5B–E) that is similar to the water structure around the U:G pair (Figure 5A) (66–69). In both cases the water structure extends to the periphery of the grooves (data not shown), with ribose 2′-hydroxyl groups serving as bridgeheads in the minor groove (70). The extensive hydration of the difluorotoluyl moiety observed in the FG structure further distinguishes the rF:G from the rF:A pair and is in good agreement with the osmotic stress data discussed above. No water molecules were observed at hydrogen-bonding distance from difluorotoluyls in rF:A pairs, although we need to keep in mind that the resolutions of the FG and FA (41) structures are different (1.1 versus 1.6 Å). Conversely, water molecules are located as close as 2.93 Å from fluorine in the FG structure (Figure 5B). If we assume that hydrogen atoms from water molecules are pointing more or less in the direction of fluorine (the resolution of the structure is not sufficient to allow visualization of H atoms), the observed OW-fluorine distances are inconsistent with mere van der Waals interactions.
Figure 6.
Stereo illustration of the superimposition of CpU-GpG [blue, NDB AR0009 (66)] and CprF-GpG (red) dinucleotide steps. The rF residue is located on the lower left.
The GA structure also revealed extensive hydration of A:G pairs in the major and minor grooves (Figure 5F) which again is in good agreement with high ▵nw obtained using osmotic stress. Our structure at higher resolution revealed the coordination of four Mg2+-hexahydrate ions inside the narrow and deep major groove. No metal cations were observed in the previously reported crystal structure of the GA dodecamer (54). Interestingly, Mg2+ ions engage in water-mediated contacts to the major groove edges of both G and A (Figure 5F).
Semi-empirical calculations of F:G and F:A pairs and their native counterparts
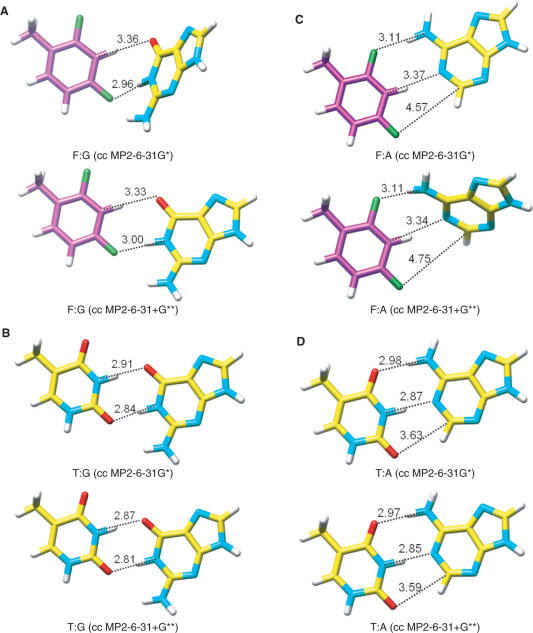
All calculations were carried out with the program GAUSSIAN (57) and a counterpoise correction (58) was applied in each case. We ran equilibrium geometry optimizations as well as point-energy calculations based on the crystallographically observed geometries. Energies were computed both at the MP2/6-31G* and MP2/6-31+G** levels of theory. These are compared in Table 3 for F:G and F:A pairs along with their native counterparts T:G and T:A. Note that we used T as the reference nucleobase due to the presence of a methyl group in 2,4-difluorotoluene. Equilibrium geometries for the various pairs obtained at both levels of theory are depicted in Figure 7. The relative ranking of energies is T:G > T:A >> F:G > F:A with energy differences between the T:G and T:A and the F:G and F:A pairs of around 3 kcal mol−1. The T:G pair is ∼8 kcal mol−1 more stable than the F:G pair (MP2/6-31+G** level). Interestingly, the order of stabilities for isolated pairs obtained from semi-empirical calculations is similar to the one established experimentally based on UV melting data for RNA duplexes containing these pairs (U instead of T; in UV melting UA is slightly more stable than UG). The geometry calculated for the F:G pair (Figure 7A) shows close resemblance to the geometries of rF:G pairs in the crystal structure of the FG duplex (Figure 5B–E). On the other hand, the calculated and experimentally observed geometries of the F:A pair differ drastically; the F4 ··· N6 distance in the calculated pair is ca. 0.7 Å shorter than the corresponding distance in the crystal structure (41). Despite this divergence between the computed and experimental pairing geometries for F:G and F:A, the differences between the average point energy based on crystallographic data and the equilibrium geometry energy for the F:G (–5.1 versus –7 kcal mol−1, respectively) and F:A pairs (–2.9 versus –4.1 kcal mol−1, respectively) are quite similar.
Table 3.
Energies (−E in kcal mol−1) obtained from semi-empirical calculations (CC = Counterpoise Correction)
| Basis set level/ Base pair | MP2 | MP2 | ||
|---|---|---|---|---|
| 6-31G* | 6-31+G** | MP2 | MP2 | |
| 6-31G* | 6-31+G** | |||
| (CC) | (CC) | |||
| Point energy calculation based on crystallographic data | ||||
| F104:G209 | 8.3 | 6.9 | 4.4 | 4.8 |
| F204:G109 | 9.2 | 7.7 | 5.1 | 5.6 |
| F304:G409 | 7.2 | 6.4 | 4.2 | 4.7 |
| F404:G309 | 8.4 | 7.1 | 4.7 | 5.1 |
| F4:A21 | 5.4 | 4.9 | 2.6 | 2.8 |
| F16: A9 | 5.6 | 4.9 | 2.6 | 2.9 |
| average F:G | 8.3 | 7 | 4.6 | 5.1 |
| average F:A | 5.5 | 4.9 | 2.6 | 2.9 |
| Energy calculation from base–pair geometry optimization | ||||
| T:A | 18.2 | 16.5 | 12.3 | 12.7 |
| F:A | 7.6 | 7.2 | 4 | 4.3 |
| T:G | 19.6 | 19.3 | 14.1 | 15.5 |
| F:G | 11.5 | 8.4 | 6.4 | 7 |
Figure 7.
Equilibrium geometries of the (A) F:G, (B) T:G, (C) F:A and (D) T:A pairs based on semi-empirical calculations [cc MP2/6-31G* and cc MP2/6-31+G** levels; cc = counterpoise correction (58)]. Distances are in Ångstrom.
DISCUSSION
Insights into Ago2 substrate recognition
In vitro siRNA experiments in HeLa cells using a dual luciferase assay demonstrated that unlike a single rF:A pair, an rF:G pair at or adjacent to the cleavage site is not tolerated by the Ago2 slicer enzyme. Similarly, a single U:G mismatch drastically reduces activity and the only mismatch pair exhibiting activity (somewhat reduced relative to a canonical siRNA duplex) is the A:C pair (41). This is consistent with a requirement for a Watson–Crick geometry of base pairs flanking the cleavage site. It is unlikely that the observed differences in activity between siRNAs containing mismatches or the rF residue at various locations are related to other factors, i.e. efficiencies in siRNA strand dissociation and biases in strand loading. Importantly, there is no correlation between the thermodynamic stability and silencing activity of the investigated siRNAs (Figure 2A, Table 1). Moreover, detailed positional analyses demonstrated that a siRNA with the rF residue in the guide strand, adjacent to the Ago2 cleavage site led to reduced silencing activity relative to positions further removed from the cleavage site or at the 5′-terminus (41,71). rF in the sense strand of the siRNA had no effect on the activity (71). However, an earlier RACE (rapid amplification of cDNA ends) analysis of the siRNA-induced cleavage of luciferase mRNA provides the strongest support that the observed silencing activities are directly related to Ago2-induced cleavage (41). Thus, cleavage at the normal site was found with siRNA AL-10 (rF:A, Figure 2A), whereas no cleavage was observed with siRNA AL-9 (rF:G, Figure 2A).
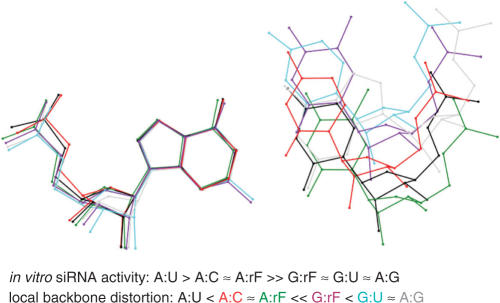
Although rF and A are more loosely paired relative to U:A, geometric constraints of the enzyme active site may produce a geometry that is not too different from that of a standard Watson–Crick base pair. Observations in connection with in vitro DNA polymerization experiments involving F-containing templates or incoming dFTP are instructive in this context (72–75). Accurate incorporation of dATP opposite template F or dFTP opposite template A is obviously not due to complementary hydrogen bond formation between the two partners (76). Rather, the active sites of high-fidelity DNA polymerases act as a mold that provides tight geometric constraints for a replicating base pair, virtually forcing it to adopt to a Watson–Crick-like arrangement (77). By contrast, the A:G, U:G and rF:G mismatches cannot be accommodated in this fashion. A superimposition of these mismatch pairs demonstrates that they result in larger deviations from a canonical backbone geometry than the rF:A and A:C pairs (Figure 8). In fact the observed siRNA activities are inversely correlated with the backbone distortions present in the RNA duplexes as a result of mismatch pairs. Although the A+:C pair with its two hydrogen bonds is expected to be isosteric with the U:G wobble pair (78), the particular example of the former used in the superimposition exhibits a smaller backbone distortion than both U:G and rF:G. Moreover, all measurements of siRNA activity were carried out at pH 7 and it is unlikely that either A or C is protonated under these conditions. A neutral A:C pair with a single hydrogen bond between N6(A) and N1(C) or N1(A) and N4(C) may in fact display a geometry that is even more similar to that of a standard Watson–Crick base pair.
Figure 8.
Comparison between the geometry of various base pairs. The diagram was generated by superimposing the purine bases (A or G, left) of different pairs. Distances between phosphates of residues on the right relative to the phosphate group of U are as follows: A:U (black, base pair A106:U207, FG structure—this article; reference structure), A:rF [green, base pair A21:rF4, 0.79 Å, (41)], G:U [cyan, AR0009, base pair G10:U5, 3.32 Å, (66)], G:rF (purple, base pair G209:rF104, 2.07 Å), A:G (gray, base pair A209:G104, GA structure—this article, 2.97 Å), and A+:C [red, AR0039, base pair A2:C17, 0.79 Å, (78)].
Contrary to the correlation between local distortions in the backbone geometry and in vitro siRNA activities, there is no obvious relationship between activity and thermodynamic stability. Thus, both the rF:A pair and the A:C pair (41) are more destabilizing than the U:G or rF:G pairs (Table 1). But the latter two lead to a loss of siRNA activity whereas the former result in a somewhat diminished activity compared to a canonical (and thermodynamically more stable) U:A pair at the Ago2 cleavage site (Figure 2). The stability data for the dodecamer duplexes with mismatches provided here and those based on longer siRNA duplexes published previously (41) are consistent with the thermodynamics at pH 7 of RNA hairpins containing single internal mismatches: G:C >> A:U >> U:G > G:A > A:C (79). Unlike siRNAs with single rF residues in place of U in the guide strand that retain activity [ref. (41) and this article], and appear to enhance sequence selectivity beyond that of the natural base in some cases (71), an siRNA with three consecutive rF:A pairs spanning the cleavage site did not display any activity (Figure 2B). This observation could indicate that multiple rF:A pairs result in more severe helical distortions (80) that are not tolerated by the Ago2 enzyme. Alternatively, the loss of activity may be related to a significant weakening of the hybridization between guide strand and mRNA target, or a combination of changes in stability and conformation.
rF:G and rF:A pairs display considerable differences
The main surprise regarding the geometry of the rF:G pair is how closely it resembles the native U:G pair, including the shear that is a hallmark of the wobble pair. Based on the significantly larger separation between rF and A relative to U and A (>0.8 Å on average based on the various atom pairs), one would perhaps have expected a looser association of rF and G as well, weakly stabilized by a C-H ··· N hydrogen bond. Instead, the average distance between atom pairs in the four independent rF:G pairs (Figure 5B–E) present in the crystal structure of the FG duplex is just 0.3 Å longer than a typical hydrogen bonding distance of ca 2.9 Å in the U:G pair (Figure 5A). Not only is the pairing tighter than expected, but the rF:G pairs display a conserved hydration pattern that matches the arrangement of water molecules around the native U:G wobble pair. By comparison, the hydration of the rF:A pair was much more limited (41). It is unlikely that this observation is simply due to the lower resolution of the earlier structure compared to that of the FG duplex (1.6 Å). Osmotic stress measurements are consistent with the structural data and support the notion that the rF:G pair is surrounded by more solvent molecules. Furthermore, the thermodynamic stability (TM) of an RNA dodecamer duplex containing two rF:G pairs is higher than that of the corresponding duplex with rF:A pairs (Table 1). This is noteworthy because U:G pairs are typically less stable than U:A pairs (79). By analogy, one would have predicted that rF:G pairs are less stable than rF:A pairs. Indeed, our findings with modified RNA duplexes differ from published stability data for DNA dodecamer duplexes with single F:G or F:A pairs (81). Accordingly, the relative order of stability in DNA is T:A (TM = 50°C) > T:G (43°C) > F:A (36°C) > F:G (35°C). Although there are no crystal structures of DNA duplexes with incorporated F:A or F:G pairs it is unlikely that the differences between DNA and RNA arise from differential stacking interactions involving the difluorotoluene moiety. Incorporation of F in place of T into DNA leads to improved stacking (82). By comparison, the particular pairing mode between F/T and G pushes the former into the major groove, thus presumably reducing the overlap with the surrounding nucleobases. Overall, X-ray crystallography, osmotic stress measurements and UV melting data all clearly indicate that F pairs more favorably with G than with A in the case of RNA.
Organic fluorine as a hydrogen-bond acceptor
The question whether fluorine can act as an acceptor of hydrogen bonds has recently been discussed in detail and with authority (65,83). In general, fluorine is a poor acceptor despite being very electronegative, and a screen of small molecule crystal structures only recovered a few examples with short X-H←d→F-Y contacts (2.1 < d < 2.3 Å). Many of these structures do not contain a substantial amount of solvent, but their precision obviously exceeds that of a macromolecular crystal structure such as that of the FG duplex even at 1.1 Å. Hydrogen atoms can typically not be located in crystal structures of macromolecules at resolutions of around 1 Å. On the other hand bond lengths and angles in such structures are sufficiently precise (r.m.s.d. bond lengths <0.02 Å, Table 2) to allow reliable statements as to the presence or absence of a hydrogen bond. In drawing this conclusion we need to be aware that a short contact involving potential acceptor and donor functions is not necessary making a crucial contribution to stability.
The crystal structure of the FG duplex reveals distances between fluorine and N-H in three of the four rF:G pairs that are consistent with formation of a hydrogen bond: The F ··· N distances range between 3.03 and 3.08 Å and, therefore, the F ··· H distances can be expected to be around 2.1 Å (Figure 5B, C and E). Interestingly, the calculated energies (Table 3; MP2 6-31+G**) based on the crystallographically observed geometries of F:G pairs correlate nicely with the F ··· N distances. Thus, the point energy of the F304:G409 pair with an F ··· N distance of 3.45 Å is –4.7 kcal/mol compared to –5.6 kcal/mol for the F204:G109 pair with a F ··· N distance of 3.03 Å. When this fluorine (F2) is replaced with a hydrogen, point energy calculations at the MP2 6-31+G** (CC) level using the geometries of the four experimentally determined F:G pairs (Figure 5, Table 3) for the 4-fluorotoluene:G pair indicate destabilizations ranging between 3.5 and 4.5 kcal mol−1. The notion that a short distance involving fluorine is associated with a stabilizing contribution certainly does not contradict the conclusion that fluorine is engaging in a hydrogen bond in F:G pairs. Further the calculations allow the conclusion that the pairing between F and G accounts for about half the stability resulting from pairing between T and G. It is unlikely, given the above observations, that pairing of F and G only involves the formation of a C-H ··· N hydrogen bond. And in line with the experimental data, the semi-empirical calculations demonstrate that the F:G pair is more stable than the F:A pair.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We are grateful for financial support by the US National Institutes of Health (grants R01 GM55237 to M.E. and R01 GM071461 to E.R.), the Austrian Science Fund FWF (P17864 to R.M.), and the BM-BWK (Gen-AU program, “Non-coding RNAs”, No. P7260-012-011, to R.M.). S.L.M. was supported by a Matz Co-op Scholarship (Northeastern University). We would like to thank Dr Zdzislaw Wawrzak for help with data collection and processing and Mr Benjamin Diop-Frimpong with assistance in the refinement of the AG RNA dodecamer. Data were collected on the 22-ID beamline of the Southeast Regional Collaborative Access Team (SER-CAT) at the Advanced Photon Source, Argonne National Laboratory. Supporting institutions may be found at www.ser-cat.org/members.html. The DuPont-Northwestern-Dow Collaborative Access Team (DND-CAT) Synchrotron Research Center at the Advanced Photon Source (Sector 5) is supported by E. I. DuPont de Nemours & Co., The Dow Chemical Company, the National Science Foundation and the State of Illinois. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under Contract No. W-31-109-Eng-38. Funding to pay the Open Access publication charges for this article was provided by grant NIH R01 GM055237.
Conflict of interest statement. None declared.
REFERENCES
- 1.Novina CD, Sharp PA. The RNAi revolution. Nature. 2004;430:161–164. doi: 10.1038/430161a. [DOI] [PubMed] [Google Scholar]
- 2.Mello CC, Conte D., Jr Revealing the world of RNA interference. Nature. 2004;431:338–342. doi: 10.1038/nature02872. [DOI] [PubMed] [Google Scholar]
- 3.Hannon GJ, Rossi JJ. Unlocking the potential of the human genome with RNA interference. Nature. 2004;431:371–378. doi: 10.1038/nature02870. [DOI] [PubMed] [Google Scholar]
- 4.Tuschl T, Borkhardt A. Small interfering RNAs: a revolutionary tool for the analysis of gene function and gene therapy. Mol. Interv. 2002;2:158–167. doi: 10.1124/mi.2.3.158. [DOI] [PubMed] [Google Scholar]
- 5.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 6.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 7.Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caplen NJ, Parrish S, Imani F, Fire A, Morgan RA. Specific inhibition of gene expression by small double-stranded RNAs in invertebrate and vertebrate systems. Proc. Natl Acad. Sci. USA. 2001;98:9742–9747. doi: 10.1073/pnas.171251798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nykanen A, Haley B, Zamore PD. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell. 2001;107:309–321. doi: 10.1016/s0092-8674(01)00547-5. [DOI] [PubMed] [Google Scholar]
- 10.Martinez J, Patkaniowska A, Urlaub H, Luhrmann R, Tuschl T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell. 2002;110:563–574. doi: 10.1016/s0092-8674(02)00908-x. [DOI] [PubMed] [Google Scholar]
- 11.Schwarz DS, Hutvagner G, Haley B, Zamore PD. Evidence that siRNAs function as guides, not primers, in the Drosophila and human RNAi pathways. Mol. Cell. 2002;10:537–548. doi: 10.1016/s1097-2765(02)00651-2. [DOI] [PubMed] [Google Scholar]
- 12.Elbashir SM, Martinez J, Patkaniowska A, Lendeckel W, Tuschl T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 2001;20:6877–6888. doi: 10.1093/emboj/20.23.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 14.Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- 15.Hutvagner G, Zamore PD. RNAi: nature abhors a double-strand. Curr. Opin. Genet. Dev. 2002;12:225–232. doi: 10.1016/s0959-437x(02)00290-3. [DOI] [PubMed] [Google Scholar]
- 16.Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 17.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 18.Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- 19.Harborth J, Elbashir SM, Vandenburgh K, Manninga H, Scaringe SA, Weber K, Tuschl T. Sequence, chemical, and structural variation of small interfering RNAs and short hairpin RNAs and the effect on mammalian gene silencing. Antisense Nucleic Acid Drug Dev. 2003;13:83–105. doi: 10.1089/108729003321629638. [DOI] [PubMed] [Google Scholar]
- 20.Holen T, Amarzguioui M, Wiiger MT, Babaie E, Prydz H. Positional effects of short interfering RNAs targeting the human coagulation trigger Tissue Factor. Nucleic Acids Res. 2002;30:1757–1766. doi: 10.1093/nar/30.8.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kretschmer-Kazemi Far R, Sczakiel G. The activity of siRNA in mammalian cells is related to structural target accessibility: a comparison with antisense oligonucleotides. Nucleic Acids Res. 2003;31:4417–4424. doi: 10.1093/nar/gkg649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gong D, Ferrell JE., Jr Picking a winner: new mechanistic insights into the design of effective siRNAs. Trends Biotechnol. 2004;22:451–454. doi: 10.1016/j.tibtech.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 23.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 24.Reynolds A, Leake D, Boese Q, Scaringe S, Marshall WS, Khvorova A. Rational siRNA design for RNA interference. Nat. Biotechnol. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- 25.Pei Y, Tuschl T. On the art of identifying effective and specific siRNAs. Nat. Methods. 2006;3:670–676. doi: 10.1038/nmeth911. [DOI] [PubMed] [Google Scholar]
- 26.Hamada M, Ohtsuka T, Kawaida R, Koizumi M, Morita K, Furukawa H, Imanishi T, Miyagishi M, Taira K. Effects on RNA interference in gene expression (RNAi) in cultured mammalian cells of mismatches and the introduction of chemical modifications at the 3'-ends of siRNAs. Antisense Nucleic Acid Drug Dev. 2002;12:301–309. doi: 10.1089/108729002761381285. [DOI] [PubMed] [Google Scholar]
- 27.Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 28.Yu JY, DeRuiter SL, Turner DL. RNA interference by expression of short-interfering RNAs and hairpin RNAs in mammalian cells. Proc. Natl Acad. Sci. USA. 2002;99:6047–6052. doi: 10.1073/pnas.092143499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paddison PJ, Caudy AA, Bernstein E, Hannon GJ, Conklin DS. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 2002;16:948–958. doi: 10.1101/gad.981002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Czauderna F, Fechtner M, Aygun H, Arnold W, Klippel A, Giese K, Kaufmann J. Functional studies of the PI(3)-kinase signalling pathway employing synthetic and expressed siRNA. Nucleic Acids Res. 2003;31:670–682. doi: 10.1093/nar/gkg141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Soutschek J, Akinc A, Bramlage B, Charisse K, Constien R, Donoghue M, Elbashir S, Geick A, Hadwiger P, et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432:173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- 32.Kurreck J. Antisense technologies. Improvement through novel chemical modifications. Eur. J. Biochem. 2003;270:1628–1644. doi: 10.1046/j.1432-1033.2003.03555.x. [DOI] [PubMed] [Google Scholar]
- 33.Manoharan M. RNA interference and chemically modified siRNAs. Nucleic Acids Res. 2003;(Suppl.3):115–116. doi: 10.1093/nass/3.1.115. [DOI] [PubMed] [Google Scholar]
- 34.Dorsett Y, Tuschl T. siRNAs: applications in functional genomics and potential as therapeutics. Nat. Rev. Drug Discov. 2004;3:318–329. doi: 10.1038/nrd1345. [DOI] [PubMed] [Google Scholar]
- 35.Manoharan M. RNA interference and chemically modified small interfering RNAs. Curr. Opin. Chem. Biol. 2004;8:570–579. doi: 10.1016/j.cbpa.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 36.Eckstein F. Small non-coding RNAs as magic bullets. Trends Biochem. Sci. 2005;30:445–452. doi: 10.1016/j.tibs.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 37.Morrissey DV, Zinnen SP, Dickinson, B.A.; Jensen K, McSwiggen JA, Vargeese C, Polisky B. Chemical modifications of synthetic siRNAs. Pharm. Discovery Dev. 2005 [Google Scholar]
- 38.Bumcrot D, Manoharan M, Koteliansky V, Sah DW. RNAi therapeutics: a potential new class of pharmaceutical drugs. Nat. Chem. Biol. 2006;2:711–719. doi: 10.1038/nchembio839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Czauderna F, Fechtner M, Dames S, Aygun H, Klippel A, Pronk GJ, Giese K, Kaufmann J. Structural variations and stabilising modifications of synthetic siRNAs in mammalian cells. Nucleic Acids Res. 2003;31:2705–2716. doi: 10.1093/nar/gkg393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kraynack BA, Baker BF. Small interfering RNAs containing full 2'-O-methylribonucleotide-modified sense strands display Argonaute2/eIF2C2-dependent activity. RNA. 2006;12:163–176. doi: 10.1261/rna.2150806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xia J, Noronha A, Toudjarska I, Li F, Akinc A, Braich R, Frank-Kamenetsky M, Rajeev KG, et al. Gene silencing activity of siRNAs with a ribo-difluorotoluyl nucleotide. ACS Chem. Biol. 2006;1:176–183. doi: 10.1021/cb600063p. [DOI] [PubMed] [Google Scholar]
- 42.Pitsch S, Weiss PA, Jenny L, Stutz A, Wu X. Reliable chemical synthesis of oligoribonucleotides (RNA) with 2’-O-[(triisopropylsylyl)oxy]methyl(2’-O-tom)-protected phosphoramidites. Helv. Chim. Acta. 2001;84:3773–3795. [Google Scholar]
- 43.Hobartner C, Micura R. Bistable secondary structures of small RNAs and their structural probing by comparative imino proton NMR spectroscopy. J. Mol. Biol. 2003;325:421–431. doi: 10.1016/s0022-2836(02)01243-3. [DOI] [PubMed] [Google Scholar]
- 44.Puglisi JD, Tinoco I., Jr Absorbance melting curves of RNA. Meth. Enzymol. 1989;180:304–325. doi: 10.1016/0076-6879(89)80108-9. [DOI] [PubMed] [Google Scholar]
- 45.Kim TW, Kool ET. A set of nonpolar thymidine nucleoside analogues with gradually increasing size. Org. Lett. 2004;6:3949–3952. doi: 10.1021/ol048487u. [DOI] [PubMed] [Google Scholar]
- 46.Breslauer KJ. Extracting thermodynamic data from equilibrium melting curves for oligonucleotide order-disorder transitions. Meth. Enzymol. 1995;259:221–242. doi: 10.1016/0076-6879(95)59046-3. [DOI] [PubMed] [Google Scholar]
- 47.Spink CH, Chaires JB. Effects of hydration, ion release, and excluded volume on the melting of triplex and duplex DNA. Biochemistry. 1999;38:496–508. doi: 10.1021/bi9820154. [DOI] [PubMed] [Google Scholar]
- 48.Rozners E, Moulder J. Hydration of short DNA, RNA and 2'-OMe oligonucleotides determined by osmotic stressing. Nucleic Acids Res. 2004;32:248–254. doi: 10.1093/nar/gkh175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Berger I, Kang CH, Sinha N, Wolters M, Rich A. A highly efficient 24-condition matrix for the crystallization of nucleic acid fragments. Acta Crystallogr. D Biol. Crystallogr. 1996;52:465–468. doi: 10.1107/s0907444995013564. [DOI] [PubMed] [Google Scholar]
- 50.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Meth. Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 51.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Cryst. 1993;26:795–800. [Google Scholar]
- 52.Kissinger CR, Gehlhaar DK, Fogel DB. Rapid automated molecular replacement by evolutionary search. Acta Crystallogr. D Biol. Crystallogr. 1999;55:484–491. doi: 10.1107/s0907444998012517. [DOI] [PubMed] [Google Scholar]
- 53.Cambillau C, Roussel A. 1997. Turbo Frodo, Version OpenGL.1. [Google Scholar]
- 54.Leonard GA, McAuley-Hecht KE, Ebel S, Lough DM, Brown T, Hunter WN. Crystal and molecular structure of r(CGCGAAUUAGCG): an RNA duplex containing two G(anti).A(anti) base pairs. Structure. 1994;2:483–494. doi: 10.1016/S0969-2126(00)00049-6. [DOI] [PubMed] [Google Scholar]
- 55.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 56.Sheldrick GM, Schneider TR. SHELXL: highresolution refinement. Meth. Enzymol. 1997;277:319–343. [PubMed] [Google Scholar]
- 57.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Montgomery JAJ, Vreven T, Kudin KN, et al. Inc., Wallingford, CT: Gaussian; 2004. edn. Revision C 02 edn. [Google Scholar]
- 58.Boys SF, Bernardi F. The calculation of small molecular interactions by the differences of separate total energies. Some procedures with reduced errors. Mol. Phys. 1970;19:553–566. [Google Scholar]
- 59.Parsegian VA, Rand RP, Rau DC. Macromolecules and water: probing with osmotic stress. Meth. Enzymol. 1995;259:43–94. doi: 10.1016/0076-6879(95)59039-0. [DOI] [PubMed] [Google Scholar]
- 60.Rozners E, Smicius R, Uchiyama C. Expanding functionality of RNA: synthesis and properties of RNA containing imidazole modified tandem G-U wobble base pairs. Chem. Commun. (Camb.) 2005:5778–5780. doi: 10.1039/b510846b. [DOI] [PubMed] [Google Scholar]
- 61.Timasheff SN. In disperse solution, “osmotic stress” is a restricted case of preferential interactions. Proc. Natl Acad. Sci. USA. 1998;95:7363–7367. doi: 10.1073/pnas.95.13.7363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Courtenay ES, Capp MW, Anderson CF, Record MT., Jr Vapor pressure osmometry studies of osmolyte-protein interactions: implications for the action of osmoprotectants in vivo and for the interpretation of "osmotic stress" experiments in vitro. Biochemistry. 2000;39:4455–4471. doi: 10.1021/bi992887l. [DOI] [PubMed] [Google Scholar]
- 63.Lavery R, Sklenar H. Defining the structure of irregular nucleic acids: conventions and principles. J. Biomol. Struct. Dyn. 1989;6:655–667. doi: 10.1080/07391102.1989.10507728. [DOI] [PubMed] [Google Scholar]
- 64.Pauling L. The Nature of the Chemical Bond. Ithaca, New York: 2nd edn. Cornell University Press; 1939. [Google Scholar]
- 65.Dunitz JD, Schweizer WB. Molecular pair analysis: C-H.F interactions in the crystal structure of fluorobenzene? And related matters. Chem. Eur. J. 2006;12:6804–6815. doi: 10.1002/chem.200600092. [DOI] [PubMed] [Google Scholar]
- 66.Mueller U, Schubel H, Sprinzl M, Heinemann U. Crystal structure of acceptor stem of tRNA(Ala) from Escherichia coli shows unique G.U wobble base pair at 1.16 A resolution. RNA. 1999;5:670–677. doi: 10.1017/s1355838299982304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Auffinger P, Westhof E. Hydration of RNA base pairs. J. Biomol. Struct. Dyn. 1998;16:693–707. doi: 10.1080/07391102.1998.10508281. [DOI] [PubMed] [Google Scholar]
- 68.Masquida B, Westhof E. On the wobble GoU and related pairs. RNA. 2000;6:9–15. doi: 10.1017/s1355838200992082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Westhof E, Fritsch V. RNA folding: beyond Watson-Crick pairs. Structure. 2000;8:R55–R65. doi: 10.1016/s0969-2126(00)00112-x. [DOI] [PubMed] [Google Scholar]
- 70.Egli M, Portmann S, Usman N. RNA hydration: a detailed look. Biochemistry. 1996;35:8489–8494. doi: 10.1021/bi9607214. [DOI] [PubMed] [Google Scholar]
- 71.Somoza A, Chelliserrykattil J, Kool ET. The roles of hydrogen bonding and sterics in RNA interference. Angew. Chem. Int. Ed. Engl. 2006;45:4994–4997. doi: 10.1002/anie.200601311. [DOI] [PubMed] [Google Scholar]
- 72.Moran S, Ren RX, Kool ET. A thymidine triphosphate shape analog lacking Watson-Crick pairing ability is replicated with high sequence selectivity. Proc. Natl Acad. Sci. USA. 1997;94:10506–10511. doi: 10.1073/pnas.94.20.10506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Moran S, Ren RX-F, Rumney S, Kool ET. Difluorotoluene, a nonpolar isostere of thymine, codes specifically and efficiently for adenine in DNA replication. J. Am. Chem. Soc. 1997;119:2056–2057. doi: 10.1021/ja963718g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Morales JC, Kool ET. Efficient replication between non-hydrogen-bonded nucleoside shape analogs. Nat. Struct. Biol. 1998;5:950–954. doi: 10.1038/2925. [DOI] [PubMed] [Google Scholar]
- 75.Delaney JC, Henderson PT, Helquist SA, Morales JC, Essigmann JM, Kool ET. High-fidelity in vivo replication of DNA base shape mimics without Watson-Crick hydrogen bonds. Proc. Natl Acad. Sci. USA. 2003;100:4469–4473. doi: 10.1073/pnas.0837277100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang X, Houk KN. Difluorotoluene, a thymine isostere, does not hydrogen bond after all. Chem. Comm. (Camb.) 1998:2631–2632. [Google Scholar]
- 77.Kim TW, Brieba LG, Ellenberger T, Kool ET. Functional evidence for a small and rigid active site in a high fidelity DNA polymerase: probing T7 DNA polymerase with variably sized base pairs. J. Biol. Chem. 2006;281:2289–2295. doi: 10.1074/jbc.M510744200. [DOI] [PubMed] [Google Scholar]
- 78.Lima S, Hildenbrand J, Korostelev A, Hattman S, Li H. Crystal structure of an RNA helix recognized by a zinc-finger protein: an 18-bp duplex at 1.6 A resolution. RNA. 2002;8:924–932. doi: 10.1017/s1355838202028893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Meroueh M, Chow CS. Thermodynamics of RNA hairpins containing single internal mismatches. Nucleic Acids Res. 1999;27:1118–1125. doi: 10.1093/nar/27.4.1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zacharias M, Engels JW. Influence of a fluorobenzene nucleobase analogue on the conformational flexibility of RNA studied by molecular dynamics simulations. Nucleic Acids Res. 2004;32:6304–6311. doi: 10.1093/nar/gkh971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Guckian KM, Krugh TR, Kool ET. Solution structure of a DNA duplex containing a replicable difluorotoluene-adenine pair. Nat. Struct. Biol. 1998;5:954–959. doi: 10.1038/2930. [DOI] [PubMed] [Google Scholar]
- 82.Guckian KM, Schweitzer BA, Ren RX, Sheils C, Paris PL, Tahmassebi DC, Kool ET. Experimental measurement of aromatic stacking affinities in the context of duplex DNA. J. Am. Chem. Soc. 1996;118:8182–8183. doi: 10.1021/ja961733f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dunitz JD. Organic fluorine: odd man out. Chembiochem. 2004;5:614–621. doi: 10.1002/cbic.200300801. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.