Abstract
Auxiliary splicing signals play a major role in the regulation of constitutive and alternative pre-mRNA splicing, but their relative importance in selection of mutation-induced cryptic or de novo splice sites is poorly understood. Here, we show that exonic sequences between authentic and aberrant splice sites that were activated by splice-site mutations in human disease genes have lower frequencies of splicing enhancers and higher frequencies of splicing silencers than average exons. Conversely, sequences between authentic and intronic aberrant splice sites have more enhancers and less silencers than average introns. Exons that were skipped as a result of splice-site mutations were smaller, had lower SF2/ASF motif scores, a decreased availability of decoy splice sites and a higher density of silencers than exons in which splice-site mutation activated cryptic splice sites. These four variables were the strongest predictors of the two aberrant splicing events in a logistic regression model. Elimination or weakening of predicted silencers in two reporters consistently promoted use of intron-proximal splice sites if these elements were maintained at their original positions, with their modular combinations producing expected modification of splicing. Together, these results show the existence of a gradient in exon and intron definition at the level of pre-mRNA splicing and provide a basis for the development of computational tools that predict aberrant splicing outcomes.
INTRODUCTION
Removal of introns from pre-messenger RNA (pre-mRNA) by splicing is a critical step in eukaryotic gene expression. Splicing of human pre-mRNAs is mediated by conserved but highly degenerate sequences at the splice sites. These signals include the YAG|R consensus (Y is pyrimidine, R is purine, | is the intron/exon boundary) at the 3′ splice site (3′ss), with upstream poly-Y tracts (PPTs) and the branch point sequence (BPS), and the MAG|GURAGU consensus (M is A or C) at the 5′ splice site (5′ss). In addition to these essential sequences, accurate recognition of exons and introns by the spliceosome requires auxiliary signals that repress or promote splicing, termed exonic or intronic splicing silencers (ESSs/ISSs) or enhancers (ESEs/ISEs). These elements are thought to act as binding sites for splicing factors, which comprise serine/arginine-rich (SR) proteins (1) or heterogeneous nuclear ribonucleoproteins (hnRNPs) (2–5). Apart from direct contacts with trans-acting factors, ESSs, ISSs, ESEs and ISEs may influence splicing through alterations of the RNA secondary structure by modifying their access to the pre-mRNA or their interactions with each other (6).
Auxiliary splicing sequences have been characterized by in vivo or in vitro selection methods (1,7–10) or through disease-associated mutations or variants that disrupted pre-mRNA splicing (11,12). In addition to experimental approaches, these elements have been identified ab initio by comparing oligomer frequencies between exons and introns and between exons with weak and strong splice sites (hexamer RESCUE-ESEs) (13), between non-coding exons and pseudoexons plus the 5′ untranslated region (UTR) of intron-less genes (putative octamer ESEs/ESSs or PESEs/PESSs) (14,15), or by evaluating conservation levels of exonic wobble positions (16). However, the relative importance of these signals in selection of 5′ or 3′ss and regulation of alternative splicing has been poorly understood.
Naturally occurring mutations or DNA variants that affect pre-mRNA splicing represent a substantial proportion of gene alterations leading to Mendelian disorders (17,18) and are thought to contribute significantly to predisposition to or protection against multifactorial or complex traits (19,20). Disease-associated splicing mutations usually lead to skipping of one or more exons or to activation of cryptic (if the mutation is in the splice-site consensus) or de novo (if the mutation is elsewhere) splice sites (21–25). Activation of cryptic splice sites can be influenced by the balance between the intrinsic strength of aberrant splice sites and their authentic counterparts (23–27), the availability of traditional splicing signals in the vicinity of mutated splice sites (18,28), exon and intron size (29), the nature of mutation (24,25) and by disrupting or creating ESEs, ESSs, ISEs or ISSs. Although exon skipping and cryptic/de novo splice-site activation account for the vast majority of aberrantly spliced RNA products transcribed from mutated alleles, the extent to which these cis-elements contribute to activation of mutation-induced splice sites has not been systematically investigated. Moreover, comparative significance of factors that determine which of the two aberrant splicing outcomes takes place is unknown.
In the present study, we have examined the frequency of computationally predicted ESEs and ESSs in a comprehensive set of 518 human sequences located between mutation-induced aberrant splice sites and their authentic counterparts. In addition, we have ascertained sequences of 250 exons that were excluded from mature transcripts as a result of disease-associated splice-site mutations and sequences of mutation-induced pseudoexons that were activated de novo in introns. We have compared these sequences to a set of controls that included intronic sequences that were never spliced, conserved exons, alternatively spliced exons and non-coding exons. In addition, we have compared sequences of the skipped exons with exons that sustained aberrant splice-site activation in vivo to identify factors that best predict the two pathological splicing outcomes. Furthermore, we have used two splicing reporter systems to experimentally test whether predicted ESSs between competing 3′ss consistently repress usage of intron-proximal sites and how their modular combinations influence 3′ss selection. Finally, using the human proinsulin gene as a model, we show how ESS mutations capable of reducing or increasing canonical transcripts alter the expression of the resulting peptide.
MATERIALS AND METHODS
Ascertainment of nucleotide sequences
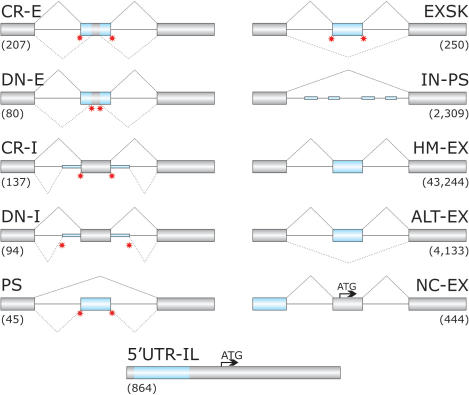
Sequences between mutation-induced aberrant 3′ss and their authentic counterparts were obtained from the updated version of DBASS3, the database of aberrant 3′ss in human disease genes (24). Sequences between aberrant 5′ss and their authentic counterparts were acquired from the recently published sister database of mutation-induced aberrant 5′ss, termed DBASS5 (25). These sequences and their annotations are available from the DBASS5 and DBASS3 web site at http://www.dbass.org.uk. Aberrant splice sites were both in exons (E) and introns (I) and were generated by mutations within (cryptic, CR) or outside (de novo, DN) 5′ or 3′ss consensus sequences in 264 human disease genes, leading to ∼250 recognizable phenotypes. The resulting sequence categories were designated CR-E, DN-E, CR-I and DN-I (Figure 1). The total number of non-repetitive aberrant 5′ and 3′ss (including pseudoexons) was 305 and 258, respectively, with 276 located in introns and 287 in exons.
Figure 1.
Schematic representation of sequence categories. Primary transcripts are shown as exons (boxes) and introns (lines). Exonic or intronic sequences analysed in this study are shown as blue boxes or blue thick lines, respectively. Canonical and aberrant splicing events are shown above and below primary transcripts, respectively. Disease-associated splicing mutations are schematically shown as red stars. Designation of each sequence category and corresponding numbers of analysed sequences is above and below the primary transcript, respectively. Arrows denote translation initiation sites. CR-E, sequences between cryptic splice sites in exons and their authentic counterparts; DN-E, sequences between de novo splice sites in exons and their authentic counterparts; CR-I, cryptic splice sites in introns; DN-I, de novo splice sites in introns; PS, sequences of mutation-induced pseudoexons (Table S2); EXSK, sequences of exons that were skipped as a result of splice-site mutations leading to disease phenotypes (Table S1); IN-PS, intronic sequences that have strong 3′ss and 5′ss and a size of 50–250 nt, but were never used by the spliceosome (14); HM-EX, human exons homologous to mouse exons (30); ALT-EX, alternatively spliced human exons (30); NC-EX, non-coding exons lacking protein-coding information (14); 5′-UTR-IL, sequences of 5′UTR in intron-less genes (14). The total length of the sequence categories (in nucleotides, nt) was 10 383; 7862; 10 686; 6988; 6114; 29 292; 352 688; 5 817 754; 513 162; 50 187 and 245 076, respectively.
In addition to aberrant splice sites, we collected sequences of 250 human exons that were skipped as a result of disease-causing mutations in natural splice sites of 127 genes (EXSK; Figure 1 and Table S1). These sequences were obtained by searching the ‘Human Gene Mutation Database’ and ‘PubMed’ for exon skipping events reported in hereditary diseases and by comparing the information in the literature to the integrated human genome databases through the ‘Ensembl’ web site. Inclusion criteria for such exons were as follows: (i) splice-site mutations were shown to be disease-causing or pre-disposing; (ii) exon skipping was detected either in patients’ RNA samples or in splicing reporter assays following transfection of wild-type and mutated constructs into mammalian cell lines, but not in controls; (iii) aberrant transcripts were characterized by nucleotide sequencing and (iv) there was no obvious utilization of aberrant splice sites in the exon or adjacent introns.
Similarly, we analysed sequences of human pseudoexons (PS) that were included in mature transcripts as a result of intronic mutations creating de novo 3′ss (n = 13) or de novo 5′ss (n = 32; Figure 1, Table S2). All these sequences were compared to controls that included human exons homologous to murine exons (HM-EX), alternatively spliced exons (ALT-EX) (30), intronic sequences that were flanked by strong splice sites but were never spliced (‘intronic pseudoexons’, IN-PS), non-coding exons (NC-EX) and intron-less sequences in the 5′-UTR (5′-UTR-IL), as described (14). Schematic representation of these sequences and their number in each category is shown in Figure 1.
Analysis of auxiliary splicing sequences
Sequences in each group were used as input files for publicly available ESE/ESS prediction algorithms at http://genes.mit.edu/burgelab/rescue-ese/(RESCUE-ESEs) (13,31), http://genes.mit.edu/fas-ess/(fluorescence-activated screen or FAS-ESSs) (10,32) and http://cubweb.biology.columbia.edu/pesx/(PESEs and PESSs) (14,15). In addition, we have used the ESEfinder (v. 3.0) (33,34) available at http://rulai.cshl.edu/tools/ESE to examine ESE motifs of four SR proteins.
The number of ESSs/ESEs in each sequence was obtained by counting each individual site that falls entirely or mostly in the sequences of mutated (sequence groups shown in the left panel of Figure 1 plus EXSK) or wild-type (right panel plus 5′-UTR-IL) alleles. Specifically, input sequences between cryptic and authentic splice site had an extra three (RESCUE-ESEs and FAS-ESSs) or four (octamer PESEs/PESSs) flanking nucleotides. To obtain the ESS/ESE density, the total number of predicted ESSs/ESEs was divided by the sequence length (in nucleotides) that excluded 3- or 4-nt overhangs and this figure was multiplied by 100. For comparison of EXSK and CR-E exons and logistic regression analysis, input exon sequences were devoid of flanking nucleotides to minimize possible confounding effects from 3′ and 5′ss signal sequences. Similarly, to determine the density of ESE motifs for SF2/ASF, SC35, SRp40 and SRp55 and their score densities, we divided their number and the sum of their scores in each sequence by its length in nucleotides and multiplied by 100. Threshold values for the original SF2/ASF (34), the updated (IgM-BRCA1) SF2/ASF version (33), SC35, SRp40 and SRp55 ESE motifs were 1.956, 1.867, 2.383, 2.67 and 2.676, respectively. The ESE motifs with scores above these standard thresholds are considered to be significant (33,34).
Splice-site prediction
The number of decoy 3′ and 5′ss in EXSK and CR-E exons was determined using the machine-learning neural network (NN) model as described (35). We used the NN splice-site predictor (v. 0.9) available at http://www.fruitfly.org/seq_tools/splice.html. To obtain the NN score density, we calculated the total number of predicted splice sites with NN scores of 0.001 and higher, divided this figure by the sequence length in nucleotides and multiplied by 100. The value of 0.001 was arbitrary to increase the power of our comparison, because many aberrant splice sites with NN scores 0.4 (default) or lower are efficiently used in vivo.
In addition to splice-site prediction ab initio, we used the maximum entropy (ME) model (36) to estimate the intrinsic strength of splice sites in most sequence categories. These scores were computed as described (24,25) using online tools available at http://genes.mit.edu/burgelab/maxent/Xmaxentscan_scoreseq_acc.html. The ME scores were shown to discriminate best aberrant 3′ (24) and 5′ss (25) from their authentic counterparts. In addition, we determined the Shapiro and Senapathy (S&S) matrix scores in EXSK and our wild-type and mutated reporter constructs. The S&S scores are based on the nucleotide frequency matrix at 3′ and 5′ss and assume independence between individual positions of splice-site consensus sequences (37,38). The S&S scores were computed using an online tool available at http://ast.bioinfo.tau.ac.il/SpliceSiteFrame.htm.
Statistical analysis
To model the relationship between predictor variables and the dichotomous splicing outcome (exon skipping and aberrant splice-site activation), we used step-wise and logistic regression analyses in S-PLUS (v. 7.0, Insightful Corp., USA). The S-PLUS was also employed to compare logistic regression models containing four to seven independent variables. To test whether there are significant differences in the mean ESE/ESS densities among sequence categories, we used the Kruskal–Wallis analysis of variance (ANOVA) on ranks in the same statistical package, followed by the Dunn's test, a non-parametric version of the Holm–Šidák multiple comparison test. Multiple comparisons of ESE/ESS proportions were carried out using the Marascuilo procedure implemented in XLSTAT (v. 2007.6; Addinsoft).
Splicing reporter constructs
Plasmids containing the entire gene for human proinsulin (INS) were prepared by amplifying a 1.5 kb genomic fragment with primers E1 (5′-ag ccc tcc agg aca ggc t) and R1 (5′-ttc aag ggc ttt att cca). The amplicons were cloned into the mammalian expression vector pCR3.1 (Invitrogen) and validated by sequencing. Minigenes containing exons 1 through 3 of the human gene for hepatic lipase (LIPC) were described previously (27). Mutated constructs were prepared with overlap-extension PCR as described (39). Plasmids were propagated in Escherichia coli DH5α and purified using the Wizard Plus SV Minipreps kit (Promega). Sequences of all constructs were analysed with the ESS/ESE prediction algorithms as described above.
Cell culture, transfections and detection of RNA products
Human embryonic kidney 293T cells were grown under standard conditions in RPMI1640 supplemented with 10% (v/v) fetal calf serum (Gibco BRL). Transient transfections were performed in 6- or 12-well plates using siPORT XP-1 (Ambion) according to the manufacturer's recommendations. Total RNA was isolated using TRI Reagent (Ambion) 48 h post-transfection and treated with DNase I (Ambion). The first-strand cDNA was reverse transcribed using oligo(dT)15 primers and Moloney murine virus reverse transcriptase (RT; Promega). RT-PCR products were amplified with primers directed to vector sequences and, as a validation control for the ratios of RNA products, with a combination of cDNA and vector primers as described (20). Exon inclusion levels were measured as described (40). The identity of each RNA product was confirmed by sequencing.
Measurement of proinsulin production
Proinsulin levels were measured in culture supernatants of 293T cells transfected with equimolar amounts wild-type INS and two mutated constructs, in which ESS/ESE mutations did not alter any amino acids (20). Total proinsulin was quantified by dissociation-enhanced lanthanide fluoroimmunoassay using monoclonal antibodies 3B1 and CPT3F11 as described (20).
RESULTS
Contribution of auxiliary splicing sequences to activation of mutation-induced aberrant splice sites
To characterize auxiliary signals underlying aberrant splice-site activation, we examined sequences located between mutation-induced cryptic (CR) or de novo (DN) splice sites in exons (E) and introns (I) and their authentic counterparts (24,25). In addition, we collected and analysed sequences of exons that sustained splice-site mutations and were skipped without activating any aberrant splice site (Table S1) and pseudoexons that were activated by intronic point mutations (Table S2). These sequences (designated CR-E, DN-E, CR-I, DN-I, EXSK and PS, respectively; Figure 1) were compared to controls that included human-mouse conserved exons (HM-EX), alternatively spliced exons (ALT-EX) (30), non-coding exons (NC-EX), 5′-UTR of intron-less genes (5′-UTR-IL) (14) and intronic segments that were flanked by strong splice sites but never included in mature transcripts (‘intronic pseudoexons’, IN-PS; Figure 1) (14).
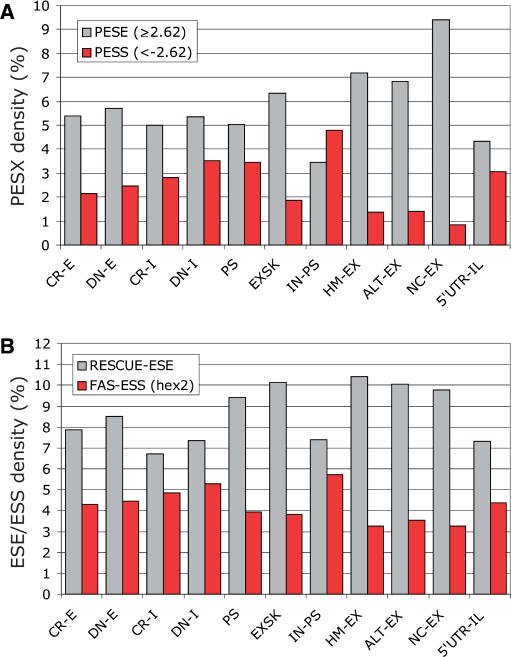
We first determined the total number of computationally predicted ESSs/ESEs in each sequence category, divided their counts by the entire nucleotide length and multiplied by 100 to obtain the ESS/ESE densities (Figures 2 and S1A). Exonic segments between cryptic/de novo splice sites and their authentic counterparts that were excluded from the mRNA as a result of splice-site mutations (CR-E and DN-E) had significantly higher PESS and FAS-ESS densities than conserved human exons (HM-EX), but lower than average introns (IN-PS; Table S3). Conversely, CR-E/DN-E segments had a lower density of PESEs and RESCUE-ESEs than HM-EX and a higher density of PESEs than IN-PS. On the other hand, newly exonized intronic segments flanking authentic splice sites (CR-I and DN-I) had a higher density of PESEs and lower PESS and FAS-ESS densities than IN-PS. More stringent sets of PESSs (the I and P scores lower than −2.88) (14) and the FAS-ESS hex3 set (10) revealed similar differences (Figure S1A). The difference in the RESCUE-ESE density between IN-PS and CR-E/DN-E or CR-I/DN-I was not strictly significant, which was partly attributable to a low number of these signals in Y-rich sequences upstream of 3′ss.
Figure 2.
Density of auxiliary splicing signals in each sequence category. Grey/red bars represent the total number of ESEs/ESSs in each sequence category divided by the total length of sequence group and multiplied by 100. Designation of sequence categories is the same as in Figure 1. (A) Density of PESE and PESS octamers (14). (B) Density of RESCUE-ESEs (13) and FAS-ESSs (32).
In addition to ESE/ESS densities, we divided ESE/ESS counts in each input sequence by its length to obtain the mean ESE/ESS densities. The differences in the median values of these densities among the sequence categories were significantly greater than expected by chance (P < 10−10 for each signal; Kruskal–Wallis ANOVA on ranks). The Dunn's test carried out with conserved exons (HMEX), exonic sequences excluded from mRNAs, exonized intronic segments and introns (Figure S2) revealed significant differences for most pair-wise comparisons, with an increasing (ESS) and decreasing (ESE) exon-to-intron gradient. Thus, the newly created exonic and intronic sequences had ESE/ESS levels intermediate between average introns and conserved exons, indicating that auxiliary signals contribute extensively to selection of mutation-induced aberrant splice sites.
Characterization of compensatory signals that promote activation of cryptic splice sites in exons
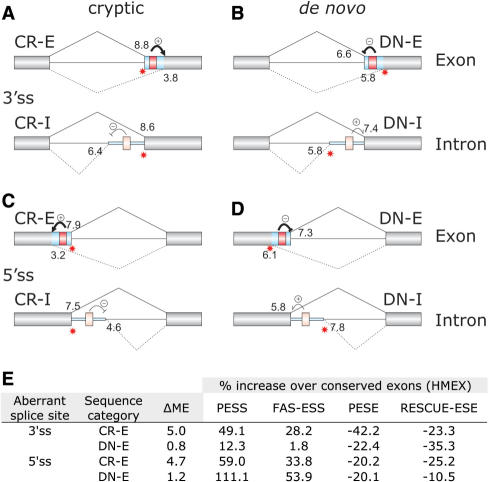
To investigate a relationship between ESEs/ESSs and traditional splice-site recognition sequences in each category of aberrant splice sites, we first determined their intrinsic strength as the average ME scores. Updated DBASS3/5 data confirmed (24,25) that cryptic splice sites in exons are exceptionally weak as compared to their authentic counterparts (Figure 3A–D), yet they were efficiently used in vivo, indicating that their activation requires auxiliary elements. To test this requirement in more detail, we compared the increase or decrease of each ESS/ESE density over HMEX in CR-E and DN-E (Figure 3E). The two categories show a marked difference (ΔME) in the mean ME scores between authentic and aberrant splice sites in exons. For aberrant 3′ss, higher ΔMEs in CR-E were accompanied by a greater increase of the FAS-ESS and PESS densities and a greater decrease in the PESE density in CR-E than in DN-E. In contrast, intrinsically weaker cryptic 5′ss had a greater decrease of the RESCUE-ESE density than de novo 5′ss. This suggests that activation of cryptic 3′ and 5′ss in exons is facilitated by differential requirements for ESE/ESS signals, with FAS-ESSs and PESXs promoting more cryptic 3′ss and RESCUE-ESEs cryptic 5′ss. However, the compensatory increase or decrease of their levels over conserved exons cannot alone explain the intrinsic weakness of CR-E or their high ΔMEs (Figure 3A, C and D), indicating that activation of cryptic splice sites in exons, particularly 5′ss, requires additional help.
Figure 3.
Predicted effect of ESSs on selection of cryptic and de novo splice sites in each sequence category. Designation of sequence categories and schematic representation of exons, introns and aberrant transcripts is the same as in Figure 1. Putative strong (red) or weak (pink) ESSs are schematically shown as boxes between cryptic (A,C) or de novo (B,D) splice sites and their authentic counterparts. Location of aberrant splice sites is shown on the right. Their expected inhibitory or stimulatory effects on competing splice sites are shown by the minus and plus signs, with weak and strong effects denoted by thin and thick arrows, respectively. The mean maximum entropy (ME) scores (36) for canonical and aberrant splice sites in each category are shown above and below the primary transcript, respectively. The updated ME scores were computed for 305 aberrant 3′ss (A,B) and 258 aberrant 5′ss (C,D) and their authentic counterparts as described (24,25). (E) Comparison of the intrinsic strength of aberrant splice sites in exons with the ESS/ESE densities. ΔME is the difference in the mean ME scores between authentic and aberrant splice sites. The mean scores are shown in panels A–D. The increase [PESS (I and P score <−2.62) and FAS-ESS (hex 2 set)] or decrease [PESE (I and P score ≥ 2.62) and RESCUE-ESE] of ESE/ESS densities in CR-E/DN-E segments over conserved exons is shown as a percentage for each sequence category.
SR ESE motif frequencies in new exonic and intronic segments
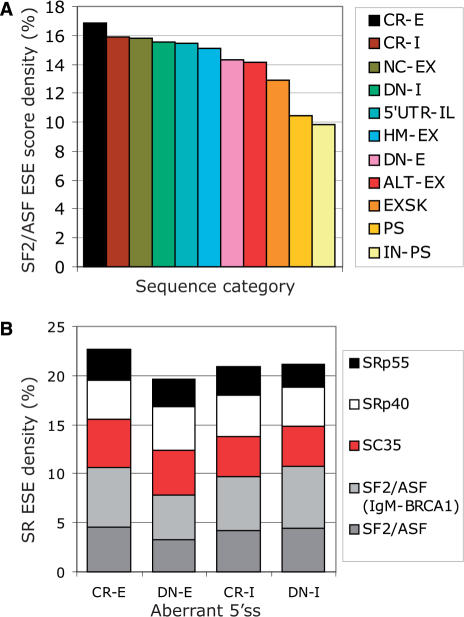
To test whether the observed differences in the ESE densities are reflected in alterations of putative ESE motifs for SR proteins (SR ESEs), we used the ESEfinder (34) to determine the SR ESE densities for SF2/ASF, SC35, SRp40 and SRp55 and their average scores in each sequence group. Of the four SR proteins, the updated score matrix of SF2/ASF (33) showed the largest spread across sequence categories (∼72%; Figure 4A), ranging from the lowest values in average introns (IN-PS), via newly included (PS) or excluded (EXSK) sequences from mRNAs, via weakly spliced (ALT-EX), conserved (HM-EX) and non-coding (NC-EX) exons and, ultimately, to pre-existing exons that were extended (CR-I) or contracted (CR-E) by mutation. Multiple comparisons of the ASF/SF2 ESE densities showed significant differences between IN-PS and both CR-E/CR-I and CR-E/DN-I, as well as among other groups (Table S4). Non-parametric ANOVA with mean SF2/ASF ESE score densities followed by the Dunn's test also discriminated segments excluded from mRNAs from both conserved exons and introns (data not shown). Importantly, the highest SF2/ASF score density was observed in CR-E segments (Figure 4A), indicating that activation of exonic cryptic splice sites relies on significant SF2/ASF help. The greatest difference in the ordered SF2/ASF ESE score densities for adjacent sequence groups was between loss of the entire exon (EXSK) and gain of the entire exon (PS) (Figure 4A). CR-E showed the highest density also for SC35 and SRp55, but not for SRp40, suggesting that SC35 and SRp55 promote selection of cryptic splice sites in exons to a lesser extent (data not shown). SRp40 had the smallest fluctuations (∼19%) among the sequence categories.
Figure 4.
Control of aberrant splice-site activation by predicted SF2/ASF ESE motifs. (A) A rainbow gradient of SF2/ASF-mediated exon/intron definition. The SF2/ASF ESE score density (IgM-BRCA1 version) was calculated as a sum of SF2/ASF ESE scores in each sequence group, divided by the sequence length and multiplied by 100. Each category is denoted by a colour shown on the right. Sequence categories were ordered from the highest score density to the lowest. For HM-EX and ALT-EX, only 1000 randomly selected sequences were analysed due to size limitations of the server. (B) The SR ESE motif densities in aberrant 5′ss. Each SR protein is represented by a colour shown on the right side.
The SR ESE density was significantly higher in 5′ss CR-E sequences than in 5′ss DN-E segments (P = 0.04). This difference was similar for the SR ESE score densities (data not shown) and was mainly due to SF2/ASF ESEs (P = 0.002 for the original SF2/ASF score matrix and 0.004 for the IgM-BRCA1 matrix; Figure 4B). For sequences between aberrant 3′ss and their authentic counterparts, we observed no significant differences, but their number was lower and their total sequence length was smaller than corresponding categories of 5′ss (Figure 1).
ESS/ESE frequencies in mutation-induced pseudoexons are intermediate between exons and introns
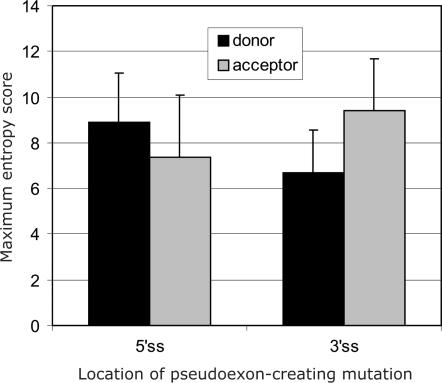
Next, we examined both traditional and auxiliary splicing sequences of cryptic exons or pseudoexons (PS; Figure 1) that were included in the mRNA as a result of intronic mutations creating de novo 5′ and 3′ss. The ME scores of splice sites created by activating mutations were significantly higher than those on the opposite ends of pseudoexons, both for mutations giving rise to the 5′ss (Wilcoxon–Mann–Whitney sum rank test; P = 0.02) and the 3′ss (P = 0.01; Figure 5). In addition to a higher PESE density in PS than in average introns, PS sequences had also an elevated RESCUE-ESE density as compared to IN-PS (Figure 2A and Table S3). Conversely, the densities of both types of silencers were lower in PS than in IN-PS. In contrast, comparison of PS and HM-EX showed a higher PESS and a lower PESE density in PS. Finally, the SF2/ASF ESE score density in PS was also intermediate between HM-EX and IN-PS (Figure 4A and Table S4). The same tendency was observed for SC35 and SRp55, with the PS levels consistently the second lowest of all sequence categories, but the second highest for SRp40 (data not shown).
Figure 5.
Pseudoexon splice sites created by intronic mutations are strong. The average ME scores of splice donor (black bars) and acceptor (grey bars) sites of pseudoexons that were activated by mutations in 5′ss (left panel) or 3′ss (right panel). Error bars represent standard deviations.
Exons that were skipped as a result of splice-site mutations are weaker than average exons
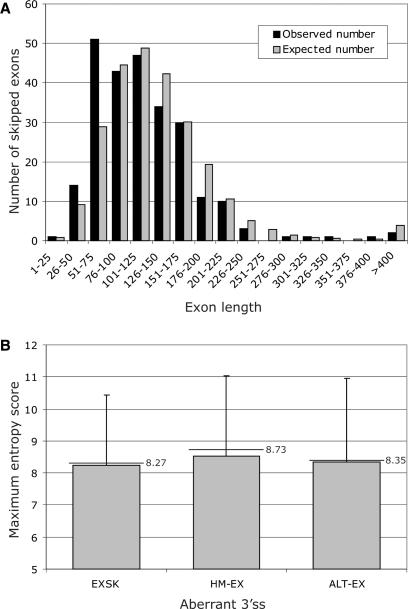
The median length of 250 exons that were skipped without activating any cryptic sites (EXSK) was significantly lower than the median size of the average exon (109 versus 122 nt, P < 0.001), but similar to a median of ALT-EX (112 nt). The most pronounced size difference between EXSK and HM-EX was observed for ∼40–75 nt exons that created a second peak in the frequency distribution (Figure 6A).
Figure 6.
Exons that were skipped as a result of splicing mutations are shorter than average exons and have weak 3′ss. (A) Length distribution of 250 skipped exons (EXSK). The observed distribution is shown as black bars. The expected distribution (grey bars) was calculated for the same number of exons using exon sizes of 43 244 homologous human-mouse exons (30). (B) Comparison of the intrinsic strength of 3′ss between exons that were skipped as a result of splice-site mutation (EXSK), and conserved (HM-EX) and alternatively spliced exons (ALT-EX). Grey bars represent means; error bars denote standard deviations; horizontal lines with labels are medians.
The mean ME scores of the wild-type EXSK 5′ss was similar to those of conserved human exons (data not shown), but the ME scores of the wild-type EXSK 3′ss were significantly lower than 3′ss of HM-EX (P = 0.04) and still somewhat lower than ALT-EX 3′ss even though most mutations leading to exon skipping were in the 5′ss (Figure 6B). Comparison of the S&S scores showed a similar tendency, but the P-value was higher (P = 0.14), most likely due to the superiority of the ME algorithm over S&S (24,25).
Because position −3 relative to the intron/exon boundary is conserved and shows a C ≥ T > A > G hierarchy in the splicing efficiency (41), we determined nucleotide frequencies at this position in introns that precede skipped exons. The frequency of As was not significantly different from that calculated for introns preceding HM-EX exons (4.8% versus 5.5%, respectively), suggesting that the differential strength of 3′ss is determined by upstream sequences, most likely by the PPT.
The PESE density was lower and the PESS density higher in EXSK than in conserved exons (Figures 2A and S1; Table S3). Although the RESCUE-ESE density was slightly lower in EXSK than in HM-EX (Figure 2B), the P value did not reach the 5% significance level in multiple comparisons (Table S3). The PESX and FAS-ESS densities for ALT-EX were intermediate between EXSK and HM-EX, with RESCUE-ESE densities in EXSK and ALT-EX virtually identical (Figure 2B).
The observed higher densities of ESSs, which are oligomers characterized by high uracil (U) content, was reflected in ∼9% U increase in EXSK over HM-EX (24.73% versus 22.77%, respectively; Figure S1B). Differences in other nucleotides between the two groups were much smaller, including As, which are markedly increased in RESCUE-ESEs as compared to background sequences of RefSeq pre-mRNAs, and Gs, which are enriched in FAS-ESSs (Figure S1B).
Dissection of factors that determine mutation-induced exon skipping and cryptic splice-site activation
To begin to disentangle the relation between multiple factors that determine the two aberrant splicing outcomes and to develop an initial predictive model, we compared sequences of exons, in which splice-site mutations activated cryptic splice sites (CR-E exons), with exons in which splice-site mutations led to exon skipping (EXSK; Table 1). Apart from a larger size of CR-E exons (P < 0.001; Wilcoxon–Mann–Whitney sum rank test), the CR-E exons had a significantly higher NN score density of predicted splice sites (P = 0.001). The NN score density of predicted 5′ss contributed more to this difference than predicted 3′ss (P = 0.002 and P = 0.07, respectively). Importantly, the PESS density in CR-E exons was significantly lower than in EXSK in multiple comparisons while the PESE and RESCUE ESE densities showed only small variations (Table 1). The CR-E exons had also a significantly higher number of SR SF2/ASF ESEs and a higher SF2/ASF score densities than EXSK (Table 1).
Table 1.
Comparison of exonic sequences that were excluded from mRNAs owing to mutations activating (CR-E exons) or failing to activate (EXSK) cryptic splice sites
| Sequence category | CR-E exonsa | EXSK | CR-E exons/EXSK |
|---|---|---|---|
| Median (mean) length in nucleotides | 148 (164.8) | 109.5 (117.2) | 1.35 (1.41) |
| Number of predicted splice sites with NN scores >0.001 (per 100 nt) | 998 (3.50) | 881 (3.01) | 1.16 |
| Sum of NN scores in predicted splice sites (per 100 nt) | 142.6 (0.50) | 104.4 (0.36) | 1.39 |
| Number of PESSs with I and P scores less than −2.62 (per 100 nt) | 313 (1.10) | 516 (1.76) | 0.63 |
| Number of PESEs with I and P scores over 2.62 (per 100 nt) | 1882 (6.60) | 1775 (6.06) | 1.09 |
| Number of PESSs with I and P scores less than −2.88 (per 100 nt) | 203 (0.71) | 342 (1.17) | 0.61 |
| Number of PESEs with I and P scores over 2.88 (per 100 nt) | 1502 (5.27) | 1423 (4.86) | 1.08 |
| Number of hex2 FAS-ESSs (per 100 nt) | 907 (3.18) | 960 (3.28) | 1.03 |
| Number of hex3 FAS-ESSs (per 100 nt) | 531 (1.86) | 534 (1.82) | 0.98 |
| Number of RESCUE-ESEs (per 100 nt) | 2633 (9.23) | 2851 (9.73) | 0.95 |
| Number of SF2/ASF ESEs (per 100 nt) | 1204 (4.22) | 957 (3.27) | 1.30 |
| Number of SF2/ASF (IgM-BRCA1) ESEs (per 100 nt) | 1698 (5.96) | 1366 (4.66) | 1.28 |
| The average SF2/ASF ESE score per 100 nt | 12.67 | 9.66 | 1.32 |
| The average SF2/ASF (IgM-BRCA1) ESEs per 100 nt | 16.92 | 12.92 | 1.31 |
| Number of SC35 ESEs (per 100 nt) | 1254 (4.40) | 1198 (4.09) | 1.08 |
| Number of SRp40 ESEs (per 100 nt) | 1203 (4.22) | 1211 (4.13) | 1.02 |
| Number of SRp55 ESEs (per 100 nt) | 732 (2.57) | 689 (2.35) | 1.09 |
| Number of all SR ESEs (per 100 nt) | 6091 (21.36) | 5421 (18.51) | 1.15 |
| Sum of all SR ESE scores (per 100 nt) | 19 440.2 (68.18) | 17 240.8 (58.86) | 1.16 |
aCR-E exonic sequences were obtained by extending the CR-E sequences [Figure 1 and ref. (24,25)] to cover the whole exon. Each exon was manually matched to the Ensembl exon–intron structure (www.ensembl.org). Only non-repetitive CR-E exons were analysed, totalling to 28 512 nucleotides. All input CR-E exon and EXSK sequences were devoid of flanking nucleotides to minimize possible confounding effects from 3′ and 5′ss signal sequences.
Unlike the RESCUE-ESE density, the SF2/ASF ESE density in CR-E was higher than in the average exon (Figures 2 and 4A), pointing to a potentially important dichotomy of RESCUE-ESEs and SR ESEs as these elements do not overlap with each other beyond what is expected by chance (42). In multiple comparisons, the entire CR-E exons had significantly higher PESE and RESCUE-ESE densities and lower PESS and FAS-ESS than CR-E segments. However, unlike the CR-E segments, the ESS densities in the CR-E exons and HM-EX were similar, with the ESE densities only marginally lower in CR-E exons (Figure 2 and Table 1, and data not shown). This strongly suggests that it is largely the sequence between authentic and aberrant splice sites that determines whether the splicing outcome is exon skipping or aberrant splice-site activation.
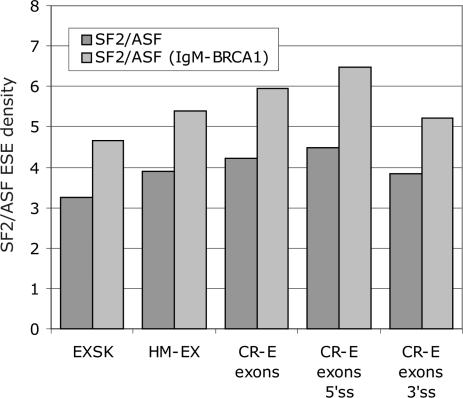
Comparison of SF2/ASF ESE densities in CR-E exons with HM-EX and EXSK showed that CR-E exons had the highest density, followed by HM-EX and EXSK (Figure 7). As with CR-E and DN-E segments (Figure 4B), the SF2/ASF ESE density was significantly higher in CR-E exons containing cryptic 5′ than 3′ss, with the latter exhibiting levels similar to conserved exons, but still somewhat higher than EXSK. Thus, the increase of predicted functional SF2/ASF sites in exons with cryptic splice sites over conserved exons is driven by 5′ss.
Figure 7.
The SF2/ASF ESE density in exons activating cryptic 5′ and 3′ss, in skipped exons and conserved human exons. Dark and light grey bars denote the total number of original SF2/ASF and updated SF2/ASF ESE motifs per 100 nt, respectively.
To model the relationship between predictor variables shown in Table 1 and the two aberrant splicing outcomes, we used multiple logistic regression with the mean ESE/ESS/SR ESE densities and their scores, exon length and the mean numbers of predicted splice sites and means of their NN scores. Binomial maximum likelihood estimates showed that the increased exon length was the best predictor of aberrant splice-site activation, followed by the mean SF2/ASF ESE score density, the density of NN scores for 5′ss and the mean PESS density (Table 2). Combining PESE and RESCUE-ESE densities into a single predictor variable did not reach significant P-values in regression analyses. Finally, employing the density of NN scores for both 3′ and 5′ss and the combined density of FAS-ESS and PESS, exon length was still the best predictor variable, followed by the SF2/ASF ESE score density, the combined 3′ and 5′ NN score density and the combined FAS-ESS and PESS density.
Table 2.
Multiple regression model to predict aberrant splice-site activation and exon skipping
| Predictor variable | Regression coefficient | Standard error | t-value | P-value |
|---|---|---|---|---|
| Exon length | 0.0092 | 0.0017 | 5.36 | <0.0001 |
| SF2/ASF ESE score densitya | 0.0535 | 0.0179 | 2.99 | <0.005 |
| The NN score density (5′ss) | 0.9679 | 0.3258 | 2.97 | <0.005 |
| PESS density | 0.1365 | 0.0673 | 2.03 | <0.05 |
Input data for logistic regression summarized in Table 1 were analysed using a generalized linear model with a logistic link (S-PLUS, v. 7.0).
aThe original SF2/ASF matrix was a consistently preferred variable over the updated (33) matrix in both stepwise and logistic regression analyses. Correlation of the two densities in EXSK and CR-exons was 0.77.
Consistent inhibition of intron-proximal 3′ss by predicted FAS-ESSs
A systematic introduction of FAS-ESS sequences between competing 3′ or 5′ss consistently inhibited usage of intron-proximal sites (32). This FAS-ESS function is in agreement with the observed higher FAS-ESS levels in CR-E/DN-E than in exons and with their lower levels in CR-I/DN-I than in introns (Figure 2 and Table S3). Stronger FAS-ESS in CR-E/DN-E would promote activation of cryptic splice sites and weaker FAS-ESS in CR-I/DN-I would promote activation of de novo splice sites (Figure 3A–D). However, a comparative analysis based on the conservation level of wobble positions in human and mouse orthologous exons suggested position-dependency of splicing regulatory sequences (16). To test the generality of these findings, we examined naturally occurring ESS/ESE motifs between competing 3′ss in human genes coding for proinsulin (INS; Figure 8) and hepatic lipase (LIPC; Figure 9).
Figure 8.
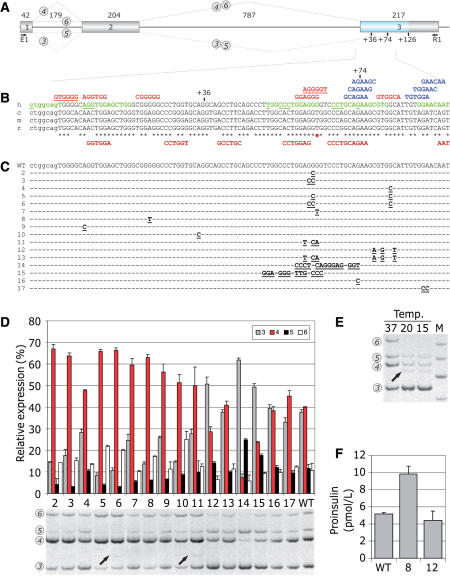
FAS-ESS-mediated inhibition of authentic 3′ss of INS intron 2 and its effects on proinsulin production. (A) INS construct. Primary transcripts are represented by exons (boxes) and introns (lines). The length of each intron and exon is shown above the primary transcript (in nucleotides). Canonical and alternative splicing is denoted by dotted lines above and below the pre-mRNA, respectively. RNA products containing exon 2 are numbered 3–6 as described previously (20). For simplicity, RNA isoforms 1 and 2 are not shown as they are expressed in very low levels. An exonic segment between two competing 3′ss is shown in blue. (B) Multiple alignment of human (h), chimpanzee (c), mouse (m) and rat (r) sequences and computationally predicted auxiliary splicing elements. Intron 2 is shown in lowercase, exon 3 is in upper case. Black asterisks denote nucleotides shared between the species. A red star shows a G/T variant in a predicted human-specific FAS-ESS. Alternative 3′ss at exon position +36 and +74 are shown by arrows. FAS-ESSs (10,32) are in red, RESCUE-ESEs (13,31) are in blue, PESEs (14) are in green and putative ESRs (16) are in brown below the sequence. The first nucleotide of octamer PESEs with the P and I scores above 2.88 (14) are underlined. The FAS-ESS hex3 set (10,32) is underlined in red. (C) Nucleotide sequences of the wild-type (WT) and mutated (2–17) splicing reporter constructs. Mutations are in bold and underlined. Nucleotides identical to the wild-type are denoted by a dash. (D) Relative expression of exon 2-containing INS mRNA isoforms. The splicing pattern of the WT and mutated (lanes 2–17) constructs is shown in the lower panel. Alternatively spliced products are shown on the left and correspond to numbers shown in panel A. Utilization (percent) of each isoform is shown in the upper panel. Error bars indicate standard deviations of a single transfection experiment in triplicate. (E) Subphysiological temperatures activate cryptic 3′ss +36 and +126 in exon 3 and promotes splicing of intron 1 in the wild-type minigene. The cryptic 3′ss (20) are denoted by arrows in panel A. Relative representation of RNA isoforms 1 and 2 that lack exon 1 was not altered (data not shown), whereas isoforms 5 and 6 were less abundant. (F) Proinsulin secretion by 293T cells following transfection of the wild-type (WT) and mutated constructs 8 and 12. Mutations are shown in panel C.
Figure 9.
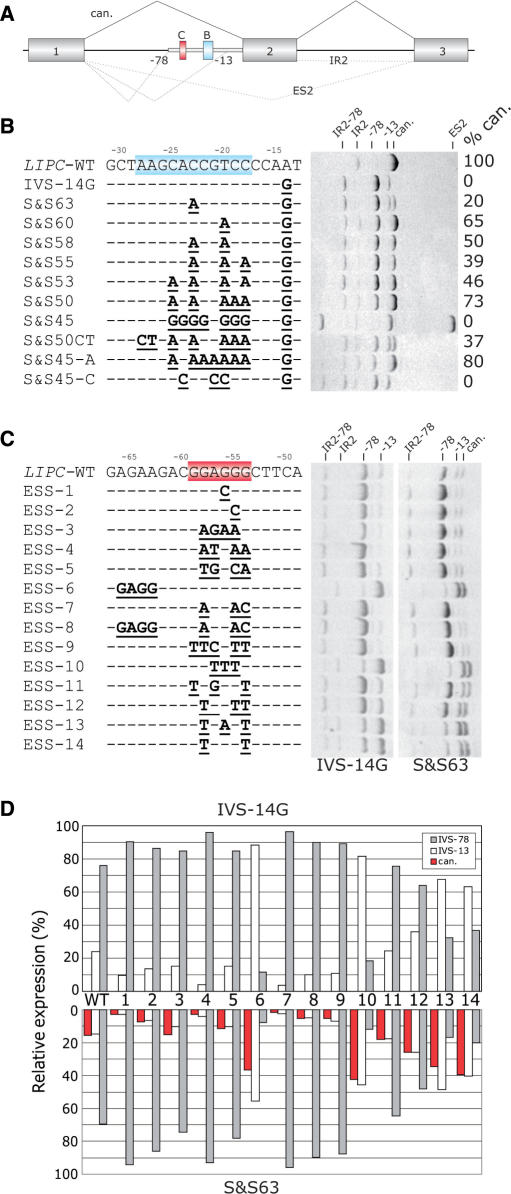
Silencer-mediated inhibition of intron-proximal 3′ss of LIPC intron 2. (A) Schematic representation of the LIPC splicing reporter. Exons, introns and aberrant splicing patterns are denoted as in Figure 8. Aberrant 3′ss 78 and 13 nt upstream of canonical (can.) 3′ss of intron 1 are designated by numbers. Blue and red boxes indicate sequences shown in panels B and C, respectively. Thick grey line represents an intronic segment retained in the mature mRNA transcribed from constructs carrying mutation IVS1-14A > G. (B) Splicing pattern of LIPC minigenes mutated upstream of the newly created 3′AG (highlighted in blue). Clone designation (S&Sn, where n is the approximate Shapiro–Senapathy score of the aberrant 3′ss 13 nt upstream of the authentic 3′ss) and mutations are shown on the left and the resulting RNA products on the right. The percentage of the canonically spliced isoform is designated with %can. The identity of each RNA product is shown at the top. IR2, retention of intron 2; ES2, skipping of exon 2. (C) Splicing pattern of LIPC minigenes mutated in a putative FAS-ESS (highlighted in red). Clone designation and mutations are shown on the left. RNA products of minigenes −14G and S&S63 (see panel B) are shown on the right. (D) Relative mRNA expression of wild-type (WT) and mutated (ESS1-14) clones lacking intron 2. Canonical products (can.) are shown in red and the amount of splicing to aberrant 3′ss −13 and −78 in white and grey, respectively.
The wild-type INS reporter activates a strong cryptic 3′ss in exon 3, 126 nt downstream of authentic 3′ss (Figure 8). We prepared a series of constructs with mutations in predicted FAS-ESSs and other motifs (Figure 8B and C), transfected mutated and wild-type plasmids into 293T cells and examined their splicing pattern (Figure 8D). Mutations were designed to avoid creation or elimination of other computationally predicted ESSs/ESEs, except for clones 11 and 13. None of the introduced changes created termination or initiation codons, nor did they create optimal splice-site consensus sequences predicted to be recognized by translation or spliceosome machineries. Clone 2, which lacked strong overlapping FAS-ESSs with a run of four Gs, showed a markedly increased utilization of canonical 3′ss. A double mutation GG > CC in clone 3, which also removed a PESE CTGGAGGG, had a similar effect. Splicing to canonical 3′ss was improved less in clone 4, in which the most distal FAS-hex2 ESS was eliminated. Clones 5 and 6 had double mutations in FAS-ESSs between competing cryptic 3′ss and also showed a significant decrease in the utilization of cryptic 3′ss. Clone 6 as well as clone 11 (see below) promoted use of a minor cryptic 3′ss 74 nt downstream of authentic 3′ss. This effect was most likely due to optimizing its PPT in the absence (clone 6) but not in the presence (clone 3) of FAS-ESS GTGGGCA, which may inhibit proximal 3′ss, or to more extensive alterations upstream of cryptic 3′ss (clone 11).
The strongest predicted FAS-ESS (GGAGGGGT) is human-specific as this motif is eliminated by a T variant present in other primates and rodents (shown by a red asterisk in Figure 8B). A G > T mutation at this position in clone 7 promoted canonical splicing. A point mutation at another non-conserved exon position (clone 8) also improved utilization of the natural 3′ss, while removing a predicted FAS-ESS containing a run of five Gs. Mutation in clone 9, which eliminated the first exonic FAS-ESS, had a similar effect. Apart from FAS-ESSs, the increased utilization of canonical 3′ss in this reporter can be explained by the removal of a competing AG dinucleotide 8 nt downstream, although this distance may be too long for efficient competition (40,43). The influence of a decoy 3′ss on 3′ss selection (3′ss +36, Figure 8B–D, 20) is illustrated by an increased canonical splicing in clone 10, in which this 3′ss was inactivated by mutation.
Interestingly, activation of a minor cryptic 3′ss +36 was not observed for any of the above mutations even after a high number of PCR cycles. Transcripts spliced to this 3′ss were found in ∼0.1% expressed sequence tags from an insulinoma library and were also detected in a pancreatic mRNA sample (20). In an attempt to induce this 3′ss, we stressed transfected 293T cells by subphysiological temperatures, which often activate aberrant splicing (44,45). Low temperatures activated the cryptic 3′ss +36 and also increased utilization of the cryptic 3′ss +126 (Figure 8E).
Combinatorial effects of FAS-ESSs on 3′ss selection
Choice of a proximal or distal splice site driven by intervening auxiliary splicing signals may reflect their position in the pre-mRNA, rather than their categorization as enhancers or silencers (16,39,46). To test whether ESSs retain their inhibitory effects on intron-proximal 3′ss when stronger silencer is replaced by a weaker one in their original positions and vice versa, we swapped two FAS-ESSs that are located in the central region between competing 3′ss (clones 11–13). In clone 11, a strong intron-proximal FAS-ESS was replaced by a weaker intron-distal FAS-ESS, resulting in duplication of the latter. Clone 12 had a duplicated version of the stronger, intron-proximal FAS-ESS instead of the intron-distal FAS-ESS and a double mutation in clone 13 exchanged positions of the two ESSs. Single mutants (clones 11 and 12) did not create or eliminate any predicted ESEs/ESSs, but clone 13 lost one natural PESE and had one extra ESR. Canonical splicing in clone 11 was increased (Figure 8D), indicating that replacement of the stronger ESS with the weaker ESS moderated inhibition of intron-proximal 3′ss. In contrast, canonical splicing was repressed in clone 12, indicating that the presence of duplicated stronger ESSs was more inhibitory for intron-proximal 3′ss than in the wild-type construct. Swapping intron-distal and intro-proximal ESSs in clone 13 had no effect. Clone 14, in which the strong overlapping FAS-ESS was moved to an adjacent position in place of an octamer PESE while removing a predicted RESCUE-ESE further downstream markedly increased splicing to the cryptic 3′ss. In clone 15, the overlapping FAS-ESSs were exchanged with an upstream PESE, also promoting cryptic 3′ss activation. Finally, single- (clone 16) or double-nucleotide (clone 17) mutations in predicted RESCUE-ESE had no or only minor effects.
As transiently transfected 293T cells are capable of secreting the gene product (20), we examined the effects of FAS-ESS mutations on the proinsulin production by measuring total proinsulin levels in culture supernatants. We employed clones 8 and 12 (Figure 8F), in which mutations did not alter amino acid sequences. Proinsulin levels were higher in clone 8 and somewhat lower in clone 12 than in the wild-type, roughly reflecting the relative expression of canonical isoform 4 (cf. Figure 8D and F), indicating that mutations in exonic splicing auxiliary signals can alter peptide production at the level of pre-mRNA splicing.
Activation of 3′ss by upstream poly(G) element associated with exon skipping
To test whether predicted auxiliary elements have similar effects on selection of an intronic cryptic 3′ss, we employed a minigene containing exons 1–3 of the human LIPC gene (Figure 9A). Here, the aberrant 3′ss was activated in the first intron 78 nt upstream of the authentic intron/exon junction and resulted from a disease-causing point mutation IVS1-14A > G that created a new AG dinucleotide downstream of the BPS (27,47). To identify cis-elements that may restore canonical splicing and to investigate utilization of competing splice sites upstream (3′ss −78) and downstream (3′ss −13) of the BPS, we first introduced a progressively increasing number of purines upstream of the newly created 3′ss. The mutant constructs had a varying strength of the 3′ss −13, with a wide range of the ME (−1.55 to −9.31) and the S&S (65.8 to 46.3) scores (Figure 9B).
The increasing number of As between position −18 to −26 correlated positively with the amount of splicing to the authentic 3′ss (r = 0.82, P < 10−15, Spearman rank order test, Figure 9B). Although As upstream of the aberrant 3′ss −13 are likely to weaken its PPT, the newly introduced As may also bring in additional consensus BPSs and generally promote 3′ss selection if located within an optimal distance from the authentic 3′ss (Figure 9B). This interpretation seems to be supported by zero utilization of authentic 3′ss following a replacement of the wild-type sequence by a stretch of Cs or Gs and activation of 3′ss −13 in clone S&S50CT, in which putative branch point As were mutated. Surprisingly, the introduction of a stretch of nine Gs upstream of the aberrant 3′ss −13 in clone S&S45 promoted use of this 3′ss while inducing significant exon skipping (Figure 9B). This mutation minimized the predicted strength of 3′ss −13 by eliminating its upstream PPT, yet it did not prevent potent activation of this splice site.
To test whether ESSs between competing 3′ss −13 and −78 consistently repress use of the intron-proximal site in a suprabranch location, we mutated minigenes IVS-14G and S&S63 in a strong predicted FAS-ESS (shown in red in Figure 9C). Point mutations in clones 1 and 2 eliminated this ESS, but did not create any predicted ESEs. Mutations in clones 3 and 4 replaced the ESS with adjacent ESEs located upstream (GAGAAG; clone 3) and downstream (GATGAA; clone 4), duplicating the ESE elements. In clone 5, the original ESS was replaced by a weaker ESS GTGGCA. Examination of the resulting splicing patterns revealed that utilization of 3′ss −78 was promoted in all clones, consistent with a relief from the FAS-ESS-mediated repression of the intron-proximal 3′ss −78. In contrast, tandem duplication of this element in clone 6 repressed the 3′ss −78 almost completely, whereas this 3′ss was strongly promoted by mutations in clone 7 that created a tandem ESE GAAGAC. Double mutations in clone 8, in which the FAS-ESS was swapped with the upstream ESE GAAGAC, promoted use of the 3′ss −78.
In clones 9–14, the ESS GGAGGG was replaced in the original position by representative FAS-ESSs from groups A, B, C, D/E, F and G (32), respectively. With the exception of clone 9, all constructs consistently promoted use of distal 3′ss, either the 3′ss −13 in constructs derived from IVS-14G or both the −13 and canonical 3′ss in mutants derived from S&S63 (Figure 9C and D). The strongest effect was observed for the FAS-ESS group B (clone 10), with a repressive hierarchy of B > F∼G > D/E > C on the 3′ss −78. Utilization of the authentic 3′ss and the 3′ss −13 was approximately equal for all mutations created in construct S&S63, consistent with the use of the same BPS for both 3′ss.
DISCUSSION
This work shows the first systematic evaluation of auxiliary splicing sequences in the development of mutation-induced aberrant splice sites. Our results clearly demonstrate that a decision to include or exclude sequences adjacent to splicing mutations in mature transcripts is influenced by their ESS/ESE frequencies (Figure 2). We show this both for exonic and intronic segments and both for cryptic and de novo splice sites. Rather than a traditionally perceived ‘binary’ concept of exon inclusion and exclusion in the mRNA, these results provide evidence for the existence of a gradient in exon and intron definition at the level of pre-mRNA splicing (Figures 2 and 4A). The observed intermediate levels of ESSs/ESEs in newly included or excluded mRNAs are consistent with recently reported intermediate ESS/ESE levels in extended segments of alternatively spliced exons (48) and high levels of alternative splicing in humans (49). They also reinforce the notion that it is the continuous exonic and intronic DNA sequence that stores information critical for correct intron removal (Figures 2 and 4A) and accurate quantitative expression of the gene product (Figure 8F). Signals carrying this information contribute significantly to a ‘splicing code’ (50), which is constituted by combinations of regulatory elements in pre-mRNAs and cellular complements of splicing factors. This code controls phenotypic consequences of splicing mutations in disease genes and locus-specific mutation patterns.
Systematic comparison of the ESS/ESE densities between exonic sequences excluded from mature transcripts and average exons revealed greater differences in the ESS densities than ESE densities, particularly in the PESS levels (Table 1, Figure 2). This finding is in agreement with recent observations of a more pronounced effect on splicing of ESSs than ESEs (28,32). The hierarchy of IN-PS > DN-I∼PS > 5′UTR-IL > CR-I > DN-E > CR-E > EXSK > ALT-EX > HM-EX > NC-EX in the PESS density was comparable to that observed for FAS-ESSs. Although the PESS density was biased towards sequences that were originally used for their selection (Figure 2A and B), these elements showed also greater differences between CR-I/DN-I and average introns than the remaining tested elements. In agreement with our data, PESSs showed the most significant overlap with auxiliary signals recently identified by a novel machine-learning algorithm (51).
Comparison of the intrinsic strength of pseudoexon 3′ and 5′ss (Figure 5) provided statistical support for a mechanism whereby a strong de novo donor or acceptor site is critical to drive the inclusion of newly recognized exons. This is consistent with a recent observation that exons with alternatively spliced 3′ or 5′ss have an intrinsically strong splice site on the fixated exon side (48) and with previous case reports (52). Once a strong anchor is introduced by mutation, exonization of intronic segments is more likely to proceed to full inclusion in the mRNA, and presumably to a more severe disease phenotype, if their ESE densities are higher and their ESS densities lower than in average introns (Figure 2). The observed second highest SRp40 ESE density in PS might suggest a distinct requirement for this protein in exon definition, in line with previous in vitro studies implicating only SRp40 (53) or only SRp40/SRp55 (54) in exon inclusion, but not the remaining SR proteins. Apart from the differential splice-site strength and ESE/ESS density, the RNA secondary structure has been recently shown to be a key feature of cryptic exon activation in the ATM and CFTR genes (55). Thus, this small but rapidly expanding sequence category may provide a useful resource for dissecting factors that influence exonization without reliance on a pre-existing splice site.
Exons that were skipped as a result of splice-site mutations were weaker than average exons, suggesting that such mutations are more likely to result in phenotypic consequences than the same mutations flanking stronger exons. The observed lower intrinsic strength of 3′ss in wild-type EXSK sequences as compared to HM-EX and ALT-EX sequences (Figure 6B) may reflect a special role of 3′ss in exon definition. Interestingly, an increase in the splice-site strength with growing intron size in evolution was more prominent for 3′ss than 5′ss (56,57). In addition to the splice-site strength, skipped exons offered both the inferior choice of decoy splice sites and the diminished chance to select them in shorter target sequences as compared to CR-E exons (Tables 1 and 2). Together with the higher SF2/ASF ESE and reduced ESS densities (Figure 2 and Tables 1 and 2), these four variables were key determinants of a decision to choose either a cryptic splice site in the exon or to opt for its exclusion from the mRNA. Although the remaining sequence-based predictor variables in logistic regression did not show significant P-values (cf. Tables 1 and 2), they are by no means excluded from future models with updated datasets. Better score matrices for the remaining SR proteins may improve the model even if their individual effects on exon inclusion and splice-site activation are smaller than SF2/ASF. Likewise, extending sequence coverage beyond the intervening segments with an enlarged DBASS3/5 sample may capture additional signals. Because the predictive value of sufficiently long individual sequences is significant (Figure S2), our results will also facilitate the development of publicly available algorithms that compute the likelihood of activating aberrant splice sites in exons as opposed to exon skipping if authentic splice sites are eliminated by mutation (I.V. et al., in preparation).
The smaller peak of the EXSK exon size distribution (Figure 6A) was largely due to frequent skipping of 54-nt exons (n = 19) of the collagen genes. These exons are rich in FAS-ESSs, but had only two PESSs (AGAATGGT, AGGATGGG). Inspection of 60 FAS-ESS hexamers identified in EXSK showed that about half of them contained GGG triplets and virtually all had two consecutive Gs, with TCCTGG, CCTGGG and GATGGG being the most frequent FAS-ESSs (Table S5). A subset of these hexamers contained optimal binding sites for hnRNP A1 (UAGGGU or UAGGGA) (4) or G-rich motifs implicated in exon silencing or intron exonization, often reflecting their relative location from authentic splice sites (39,46,58,59). Some of the collagen exons that were skipped as a result of splicing mutations had an extraordinary density of FAS-ESSs. For example, the mean FAS-ESS density of COL6A1 exon 14 (60), COL1A1 exon 49 (61) and COL3A1 exon 37 (62) was 18.52, 12.96 and 11.11, respectively, with the latter exon completely lacking RESCUE-ESEs. The intrinsic strength of 3′ and 5′ss of 54-nt exons was not significantly different from the average exon (data not shown), suggesting that their propensity to exon skipping is largely due to the small exon size and increased numbers of FAS-ESSs. These features may have contributed to the extraordinary evolution of collagen genes, which are characterized by multiple 54-nt exons encoding Gly-X1-X2 triplets in the helical domain (63). These exons were occasionally duplicated or triplicated, possibly through partially processed RNAs (63).
Of four tested SR matrices, SF2/ASF gave the best discrimination of newly intronized and exonized sequence categories and the largest spread across the exon-intron spectrum (Figure 4A and Table S4). This is consistent with the lowest P-values observed for SF2/ASF of the four proteins when comparing SR ESE motif frequencies in exons and introns (42). The elevated SF2/ASF ESE levels between cryptic 5′ss and their authentic counterparts as compared to de novo 5′ss and average exons (Figures 4B and 7) suggests that they compensate the relative lack of splicing silencers in 5′ss CR-E (Figure 3E). They may also explain a previous observation of more pronounced effects of ESEs on competing 5′ss than 3′ss (32). As SF2/ASF ESEs and high-score PESEs significantly overlap (33) and FAS-hex3 are enriched in alternative 5′ exons (10), future studies with a higher number of 5′ss CR-E sequences should determine a spatial relationship between these elements and estimate to what extent the observed 5′ to 3′ss bias can be explained by greater availability of decoy 5′ss than 3′ss in CR-E exons (Table 1). Reduced availability of 5′ss recognition sequences in the vicinity of exons that were skipped has been recently noticed for NF1 (28) and other genes (18). Nevertheless, dominant contribution of the 5′ss NN score density to differences in the overall NN score density is likely to reflect greater sequence constraints imposed by the BPS-PPT-3′YAG signals than by the less extensive 5′ss consensus.
Our experimental data confirm and extend the previous finding (32) that FAS-ESSs consistently inhibit intron-proximal splice sites. The only exception was the FAS-ESS group A (TTCGTT; Figure 9), which showed the weakest effect of all representative FAS-ESS with other reporters (32). In our experiments, the inhibitory effects of predicted FAS-ESSs on intron-proximal 3′ss were retained if these elements were maintained at their original positions in the pre-mRNA and were predictably modified by combining the auxiliary signals in a modular, possibly additive manner and by removing a decoy 3′AG nearby. Our unexpected observation of significant utilization for newly created AGs in the AG exclusion zone in the unfavourable context of a preceding stretch of Gs (Figure 9B) can be mechanistically explained by creating a secondary structure altering accessibility of spliceosomal complexes, by elimination of the authentic BPS that weakens exon definition and leads to exons skipping, by introduction of strong ESSs that completely repress the intron-proximal 3′ss −78, most likely through factors that bind to poly(G) elements, such as hnRNP H/H′/F (3,5,39,46), and activate distal the 3′ss −13, or by a combination of these factors.
In conclusion, in vivo selection of aberrant splice sites in introns and exons is extensively controlled by auxiliary splicing signals. Exonic sequences that are excluded from the mRNA as a result of mutations activating aberrant splice sites have more ESSs and less ESEs than average conserved exons. Conversely, intronic sequences that are included in the mRNAs have more ESEs and less ESSs than average introns. Second, efficient use of intrinsically weak cryptic splice sites in exons is facilitated by a higher than average density of ESSs, which promote activation of intron-distal sites, particularly cryptic 3′ss (Figures 2 and 3E), and a high density of SF2/ASF ESE motifs, especially for cryptic 5′ss (Figures 4B and 7). Third, exons that are skipped as a result of splice-site mutations are smaller than typical exons (∼90% of the median length of conserved exons), have weaker 3′ss (∼95% of the median ME score), higher than average density of predicted ESSs and lower than average density of ESEs. Fourth, a decision to exclude the whole exon or activate exonic aberrant splice site if one of the authentic splice sites is inactivated by mutation is largely driven by the overall availability of intrinsically strong decoy splice sites, SF2/ASF support and the balance between the ESS density in sequences between authentic and aberrant splice sites and the remaining exonic sequences. Fifth, predicted ESSs can promote efficient selection of 3′ss in a highly unfavourable context, highlighting the power of auxiliary sequences to control selection of many intrinsically weak splice sites in the genome. Finally, altered 3′ss selection through mutations in predicted ESSs and the resulting quantitative changes in canonical mRNAs and peptide production reinforce the notion that alternative splicing is an important but underappreciated mechanism of fine-tuning gene expression and that gene variants located in these elements are likely to contribute significantly to inter-individual phenotypic variability.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
This work was supported by the grant from the Juvenile Diabetes Research Foundation International (1-2006-263) to I.V. We thank Martin Chivers (University of Southampton) for technical assistance, Sarah Ennis and Nik Maniatis (University of Southampton) for statistical advice and Christine Glenn and Peter Wood (Department of Endocrinology, Southampton Hospital NHS Trust) for measurements of total proinsulin. Funding to pay the Open Access publication charges for this article was provided by the Juvenile Diabetes Research Foundation International.
Conflict of interest statement. None declared.
REFERENCES
- 1.Liu HX, Zhang M, Krainer AR. Identification of functional exonic splicing enhancer motifs recognized by individual SR proteins. Genes Dev. 1998;12:1998–2012. doi: 10.1101/gad.12.13.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ghetti A, Pinol-Roma S, Michael WM, Morandi C, Dreyfuss G. hnRNP I, the polypyrimidine tract-binding protein: distinct nuclear localization and association with hnRNAs. Nucleic Acids Res. 1992;20:3671–3678. doi: 10.1093/nar/20.14.3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matunis MJ, Xing J, Dreyfuss G. The hnRNP F protein: unique primary structure, nucleic acid-binding properties, and subcellular localization. Nucleic Acids Res. 1994;22:1059–1067. doi: 10.1093/nar/22.6.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burd CG, Dreyfuss G. RNA binding specificity of hnRNP A1: significance of hnRNP A1 high-affinity binding sites in pre-mRNA splicing. EMBO J. 1994;13:1197–1204. doi: 10.1002/j.1460-2075.1994.tb06369.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caputi M, Zahler AM. Determination of the RNA binding specificity of the heterogeneous nuclear ribonucleoprotein (hnRNP) H/H'/F/2H9 family. J. Biol. Chem. 2001;276:43850–43859. doi: 10.1074/jbc.M102861200. [DOI] [PubMed] [Google Scholar]
- 6.Buratti E, Baralle FE. Influence of RNA secondary structure on the pre-mRNA splicing process. Mol. Cell. Biol. 2004;24:10505–10514. doi: 10.1128/MCB.24.24.10505-10514.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coulter LR, Landree MA, Cooper TA. Identification of a new class of exonic splicing enhancers by in vivo selection. Mol. Cell. Biol. 1997;17:2143–2150. doi: 10.1128/mcb.17.4.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schaal TD, Maniatis T. Selection and characterization of pre-mRNA splicing enhancers: identification of novel SR protein-specific enhancer sequences. Mol. Cell. Biol. 1999;19:1705–1719. doi: 10.1128/mcb.19.3.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singh NN, Androphy EJ, Singh RN. In vivo selection reveals combinatorial controls that define a critical exon in the spinal muscular atrophy genes. RNA. 2004;10:1291–1305. doi: 10.1261/rna.7580704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Z, Rolish ME, Yeo G, Tung V, Mawson M, Burge CB. Systematic identification and analysis of exonic splicing silencers. Cell. 2004;119:831–845. doi: 10.1016/j.cell.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 11.Cartegni L, Chew SL, Krainer AR. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat. Rev. Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- 12.Pagani F, Stuani C, Tzetis M, Kanavakis E, Efthymiadou A, Doudounakis S, Casals T, Baralle FE. New type of disease causing mutations: the example of the composite exonic regulatory elements of splicing in CFTR exon 12. Hum. Mol. Genet. 2003;12:1111–1120. doi: 10.1093/hmg/ddg131. [DOI] [PubMed] [Google Scholar]
- 13.Fairbrother WG, Yeh RF, Sharp PA, Burge CB. Predictive identification of exonic splicing enhancers in human genes. Science. 2002;297:1007–1013. doi: 10.1126/science.1073774. [DOI] [PubMed] [Google Scholar]
- 14.Zhang XH, Chasin LA. Computational definition of sequence motifs governing constitutive exon splicing. Genes Dev. 2004;18:1241–1250. doi: 10.1101/gad.1195304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang XH, Kangsamaksin T, Chao MS, Banerjee JK, Chasin LA. Exon inclusion is dependent on predictable exonic splicing enhancers. Mol. Cell. Biol. 2005;25:7323–7332. doi: 10.1128/MCB.25.16.7323-7332.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goren A, Ram O, Amit M, Keren H, Lev-Maor G, Vig I, Pupko T, Ast G. Comparative analysis identifies exonic splicing regulatory sequences—the complex definition of enhancers and silencers. Mol. Cell. 2006;22:769–781. doi: 10.1016/j.molcel.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 17.Cooper TA, Mattox W. The regulation of splice-site selection, and its role in human disease. Am. J. Hum. Genet. 1997;61:259–266. doi: 10.1086/514856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krawczak M, Thomas NS, Hundrieser B, Mort M, Wittig M, Hampe J, Cooper DN. Single base-pair substitutions in exon-intron junctions of human genes: nature, distribution, and consequences for mRNA splicing. Hum. Mutat. 2007;28:150–158. doi: 10.1002/humu.20400. [DOI] [PubMed] [Google Scholar]
- 19.Nissim-Rafinia M, Kerem B. Splicing regulation as a potential genetic modifier. Trends Genet. 2002;18:123–127. doi: 10.1016/s0168-9525(01)02619-1. [DOI] [PubMed] [Google Scholar]
- 20.Královičová J, Gaunt TR, Rodriguez S, Wood PJ, Day INM, Vořechovský I. Variants in the human insulin gene that affect pre-mRNA splicing: is -23HphI a functional single nucleotide polymorphism at IDDM2? Diabetes. 2006;55:260–264. doi: 10.2337/diabetes.55.01.06.db05-0773. [DOI] [PubMed] [Google Scholar]
- 21.Krawczak M, Reiss J, Cooper DN. The mutational spectrum of single base-pair substitutions in mRNA splice junctions of human genes: causes and consequences. Hum. Genet. 1992;90:41–54. doi: 10.1007/BF00210743. [DOI] [PubMed] [Google Scholar]
- 22.Nakai K, Sakamoto H. Construction of a novel database containing aberrant splicing mutations of mammalian genes. Gene. 1994;141:171–177. doi: 10.1016/0378-1119(94)90567-3. [DOI] [PubMed] [Google Scholar]
- 23.Roca X, Sachidanandam R, Krainer AR. Intrinsic differences between authentic and cryptic 5′ splice sites. Nucleic Acids Res. 2003;31:6321–6333. doi: 10.1093/nar/gkg830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vořechovský I. Aberrant 3′ splice sites in human disease genes: mutation pattern, nucleotide structure and comparison of computational tools that predict their utilization. Nucleic Acids Res. 2006;34:4630–4641. doi: 10.1093/nar/gkl535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buratti E, Chivers MC, Královičová J, Romano M, Baralle M, Krainer AR, Vořechovský I. Aberrant 5′ splice sites in human disease genes: mutation pattern, nucleotide structure and comparison of computational tools that predict their utilization. Nucleic Acids Res. 2007;35:4250–4263. doi: 10.1093/nar/gkm402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roca X, Sachidanandam R, Krainer AR. Determinants of the inherent strength of human 5′ splice sites. RNA. 2005;11:683–698. doi: 10.1261/rna.2040605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Královičová J, Christensen MB, Vořechovský I. Biased exon/intron distribution of cryptic and de novo 3′ splice sites. Nucleic Acids Res. 2005;33:4882–4898. doi: 10.1093/nar/gki811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wimmer K, Roca X, Beiglbock H, Callens T, Etzler J, Rao AR, Krainer AR, Fonatsch C, Messiaen L. Extensive in silico analysis of NF1 splicing defects uncovers determinants for splicing outcome upon 5′ splice-site disruption. Hum. Mutat. 2007;28:599–612. doi: 10.1002/humu.20493. [DOI] [PubMed] [Google Scholar]
- 29.Sterner DA, Carlo T, Berget SM. Architectural limits on split genes. Proc. Natl Acad. Sci. USA. 1996;93:15081–15085. doi: 10.1073/pnas.93.26.15081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carmel I, Tal S, Vig I, Ast G. Comparative analysis detects dependencies among the 5′ splice-site positions. RNA. 2004;10:828–840. doi: 10.1261/rna.5196404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fairbrother WG, Yeo GW, Yeh R, Goldstein P, Mawson M, Sharp PA, Burge CB. RESCUE-ESE identifies candidate exonic splicing enhancers in vertebrate exons. Nucleic Acids Res. 2004;32:W187–W190. doi: 10.1093/nar/gkh393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Z, Xiao X, Van Nostrand E, Burge CB. General and specific functions of exonic splicing silencers in splicing control. Mol. Cell. 2006;23:61–70. doi: 10.1016/j.molcel.2006.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith PJ, Zhang C, Wang J, Chew SL, Zhang MQ, Krainer AR. An increased specificity score matrix for the prediction of SF2/ASF-specific exonic splicing enhancers. Hum. Mol. Genet. 2006;15:2490–2508. doi: 10.1093/hmg/ddl171. [DOI] [PubMed] [Google Scholar]
- 34.Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR. ESEfinder: a web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003;31:3568–3571. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reese MG, Eeckman FH, Kulp D, Haussler D. Improved splice site detection in Genie. J. Comput. Biol. 1997;4:311–323. doi: 10.1089/cmb.1997.4.311. [DOI] [PubMed] [Google Scholar]
- 36.Yeo G, Burge CB. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J. Comput. Biol. 2004;11:377–394. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- 37.Shapiro MB, Senapathy P. RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res. 1987;15:7155–7174. doi: 10.1093/nar/15.17.7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Senapathy P, Shapiro MB, Harris NL. Splice junctions, branch point sites, and exons: sequence statistics, identification, and applications to genome project. Methods Enzymol. 1990;183:252–278. doi: 10.1016/0076-6879(90)83018-5. [DOI] [PubMed] [Google Scholar]
- 39.Královičová J, Vořechovský I. Position-dependent repression and promotion of DQB1 intron 3 splicing by GGGG motifs. J. Immunol. 2006;176:2381–2388. doi: 10.4049/jimmunol.176.4.2381. [DOI] [PubMed] [Google Scholar]
- 40.Lei H, Královičová I. Identification of splicing silencers and enhancers in sense Alus: a role for pseudo-acceptors in splice site repression. Mol. Cell. Biol. 2005;25:6912–6920. doi: 10.1128/MCB.25.16.6912-6920.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith CW, Chu TT, Nadal-Ginard B. Scanning and competition between AGs are involved in 3′ splice site selection in mammalian introns. Mol. Cell. Biol. 1993;13:4939–4952. doi: 10.1128/mcb.13.8.4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang J, Smith PJ, Krainer AR, Zhang MQ. Distribution of SR protein exonic splicing enhancer motifs in human protein-coding genes. Nucleic Acids Res. 2005;33:5053–5062. doi: 10.1093/nar/gki810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chua K, Reed R. An upstream AG determines whether a downstream AG is selected during catalytic step II of splicing. Mol. Cell. Biol. 2001;21:1509–1514. doi: 10.1128/MCB.21.5.1509-1514.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gemignani F, Sazani P, Morcos P, Kole R. Temperature-dependent splicing of beta-globin pre-mRNA. Nucleic Acids Res. 2002;30:4592–4598. doi: 10.1093/nar/gkf607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Královičová J, Houngninou-Molango S, Kramer A, Vořechovský I. Branch site haplotypes that control alternative splicing. Hum. Mol. Genet. 2004;13:3189–3202. doi: 10.1093/hmg/ddh334. [DOI] [PubMed] [Google Scholar]
- 46.Han K, Yeo G, An P, Burge CB, Grabowski PJ. A combinatorial code for splicing silencing: UAGG and GGGG motifs. PLoS Biol. 2005;3:e158. doi: 10.1371/journal.pbio.0030158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brand K, Dugi KA, Brunzell JD, Nevin DN, Santamarina-Fojo S. A novel A > G mutation in intron 1 of the hepatic lipase gene leads to alternative splicing resulting in enzyme deficiency. J. Lipid Res. 1996;37:1213–1223. [PubMed] [Google Scholar]
- 48.Koren E, Lev-Maor G, Ast G. The emergence of alternative 3' and 5' splice site exons from constitutive exons. PLoS Comput. Biol. 2007;3:e95. doi: 10.1371/journal.pcbi.0030095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnson JM, Castle J, Garrett-Engele P, Kan Z, Loerch PM, Armour CD, Santos R, Schadt EE, Stoughton R, et al. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- 50.Matlin AJ, Clark F, Smith CW. Understanding alternative splicing: towards a cellular code. Nat. Rev. Mol. Cell. Biol. 2005;6:386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- 51.Dogan RI, Getoor L, Wilbur WJ, Mount SM. Features generated for computational splice-site prediction correspond to functional elements. BMC Bioinformatics. 2007 doi: 10.1186/1471-2105-8-410. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Buratti E, Baralle M, Baralle FE. Defective splicing, disease and therapy: searching for master checkpoints in exon definition. Nucleic Acids Res. 2006;34:3494–3510. doi: 10.1093/nar/gkl498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Du K, Peng Y, Greenbaum LE, Haber BA, Taub R. HRS/SRp40-mediated inclusion of the fibronectin EIIIB exon, a possible cause of increased EIIIB expression in proliferating liver. Mol. Cell. Biol. 1997;17:4096–4104. doi: 10.1128/mcb.17.7.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ramchatesingh J, Zahler AM, Neugebauer KM, Roth MB, Cooper TA. A subset of SR proteins activates splicing of the cardiac troponin T alternative exon by direct interactions with an exonic enhancer. Mol. Cell. Biol. 1995;15:4898–4907. doi: 10.1128/mcb.15.9.4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Buratti E, Dhir A, Lewandowska E, Baralle FE. RNA structure is a key regulatory element in pathological ATM and CFTR pseudoexon inclusion. Nucleic Acids Res. 2007;35:4369–4383. doi: 10.1093/nar/gkm447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dewey CN, Rogozin IB, Koonin EV. Compensatory relationship between splice sites and exonic splicing signals depending on the length of vertebrate introns. BMC Genomics. 2006;7:311. doi: 10.1186/1471-2164-7-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bannai H, Inenaga S, Shinohara A, Takeda M, Miyano S. A string pattern regression algorithm and its application to pattern discovery in long introns. Genome Inform. 2002;13:3–11. [PubMed] [Google Scholar]
- 58.McCullough AJ, Berget SM. G triplets located throughout a class of small vertebrate introns enforce intron borders and regulate splice site selection. Mol. Cell. Biol. 1997;17:4562–4571. doi: 10.1128/mcb.17.8.4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sironi M, Menozzi G, Riva L, Cagliani R, Comi GP, Bresolin N, Giorda R, Pozzoli U. Silencer elements as possible inhibitors of pseudoexon splicing. Nucleic Acids Res. 2004;32:1783–1791. doi: 10.1093/nar/gkh341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lamande SR, Shields KA, Kornberg AJ, Shield LK, Bateman JF. Bethlem myopathy and engineered collagen VI triple helical deletions prevent intracellular multimer assembly and protein secretion. J. Biol. Chem. 1999;274:21817–21822. doi: 10.1074/jbc.274.31.21817. [DOI] [PubMed] [Google Scholar]
- 61.Griffith AJ, Sprunger LK, Sirko-Osadsa DA, Tiller GE, Meisler MH, Warman ML. Marshall syndrome associated with a splicing defect at the COL1A1 locus. Am. J. Hum. Genet. 1998;62:816–823. doi: 10.1086/301789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schwarze U, Goldstein JA, Byers PH. Splicing defects in the COL3A1 gene: marked preference for 5′ (donor) spice-site mutations in patients with exon-skipping mutations and Ehlers-Danlos syndrome type IV. Am. J. Hum. Genet. 1997;61:1276–1286. doi: 10.1086/301641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sykes B. The molecular genetics of collagen. BioEssays. 1985;3:112–117. doi: 10.1002/bies.950030306. [DOI] [PubMed] [Google Scholar]