Abstract
To study the role of the interferon- (IFN) γR2 chain in IFN-γ signaling and immune function, IFN-γR2-deficient mice have been generated and characterized. Cells derived from IFN-γR2 −/− mice are unable to activate either JAK/STAT signaling proteins or gene transcription in response to IFN-γ. The lack of IFN-γ responsiveness alters IFN-γ-induced Ig class switching by B cells from these mice. In vitro cultures of T cells demonstrate that the T cells from the IFN-γR2 −/− mice have a defect in Th1 cell differentiation. The IFN-γR2 (−/−) mice also produce lower amounts of IFN-γ in response to antigenic challenge. In addition, IFN-γR2 −/− mice are defective in contact hypersensitivity and are highly susceptible to infection by Listeria monocytogenes. These results demonstrate that the IFN-γR2 is essential for IFN-γ-mediated immune responses in vivo.
Interferon-γ (IFN-γ) is a dimeric glycoprotein that has a variety of immune regulatory functions. Although the production of IFN-γ can be induced in several cell types (1), most IFN-γ secreted in an immune response is produced by activated T cells and natural killer (NK) cells (reviewed in refs. 2–4). The cell surface receptor complex for IFN-γ is composed of two chains, termed either IFN-γR1 and IFN-γR2 or IFN-γRα and IFN-γRβ, respectively (reviewed in refs. 4–6). The IFN-γ homodimer binds two molecules of the IFN-γR1 receptor chain. Although the IFN-γR2 chain, which is also termed accessory factor 1, cannot bind IFN-γ by itself (7), it is required for IFN-γ signaling and receptor complex formation (8–14).
Previous work has established the role of JAK and STAT molecules in IFN-γ signaling (reviewed in refs. 4 and 15). Two JAK kinases, Jak1 and Jak2, are required for the initiation of IFN-γ signaling (12, 16–19). Jak1 is constitutively associated with the IFN-γR1 while Jak2 is associated with the IFN-γR2 (12, 19). Upon addition of IFN-γ, JAKs are activated and phosphorylate the IFN-γR1 (20–22). The phosphorylated receptor recruits the cytoplasmic transcription factor Stat1 to the receptor complex. Activated JAKs are then able to phosphorylate the recruited Stat1 on tyrosine 701 (17). Two phosphorylated Stat1 molecules then homodimerize through interaction between their SH2 domains and phosphorylated tyrosine motifs. Subsequently, this dimeric Stat1 complex translocates into the nucleus to activate gene transcription (23, 24). In addition to the JAK–STAT pathway, there is evidence that several other signal transduction pathways, such as protein kinase C, sphingomyelin hydrolysis, and IRS-2, can be activated by IFN-γ (2, 25–28). However, the analysis of Stat1-deficient mice supports the concept that Stat1 is required for most of the biologic responses induced by IFN-γ (29, 30).
The ability of certain T cells to respond to IFN-γ is highly regulated through the modulated expression of the IFN-γR2 gene. Previous work has demonstrated that the IFN-γR2 gene is expressed in Th2 cells but not in Th1 cells. The lack of IFN-γR2 renders Th1 cells unresponsive to IFN-γ. In contrast, the IFN-γR1 chain is expressed at a similar level in both cell types (31–34). Interestingly, when naive T cells from the IFN-γR1-deficient mice are differentiated in vitro to Th1 cells by interleukin (IL) 12, the expression of the IFN-γR2 gene is not decreased (34). This result suggests that the down-regulation of the IFN-γR2 may not be intrinsically linked to the polarization of cells to the Th1 lineage. In contrast, Th2 populations, generated in the presence of both IL-4 and IFN-γ expressed levels of the IFN-γR2 mRNA similar to those produced by cells differentiated in the presence of IL-4 only (35). This result suggests that the IFN-γR2 down-regulation is not simply the desensitization to IFN-γ, but is coupled to the IFN-γ-dependent Th1 differentiation pathway. However, because these studies have been performed in vitro with cultured primary T cells or clones, it is not clear whether the down-regulation of IFN-γR2 is essential for Th1-mediated immune response in vivo.
To further elucidate the role of IFN-γR2 in vivo, IFN-γR2-deficient (−/−) mice were generated by gene-targeted mutagenesis. IFN-γR2 −/− mice developed normally and were fertile. Nevertheless, these mice display profound defects in IFN-γ responsiveness as measured by JAK/STAT activation in multiple cell types. Several immune responses that are induced by IFN-γ, such as Ig class switching and Th1 differentiation, are altered in these mice. In addition, these mice are susceptible to the intracellular parasite Listeria monocytogenes. These results demonstrate an essential role of IFN-γR2 in IFN-γ-mediated immune responses.
MATERIALS AND METHODS
Generation of the IFN-γR2 −/− Mutant Mouse.
The replacement vector was derived from an 11-kb EcoRI fragment isolated from a library prepared with DNA from mouse strain 129/Sv (36). The SmaI fragment containing exon 2 was replaced by a neomycin resistance cassette (phosphoglycerate kinase–neo). The targeting vector was transfected into W9.5 ES cells by electroporation, and transfectants were selected with G418 (350 μg/ml) and FIAU (50 pM) as described previously (37). Genomic DNA was cut by BamHI and blotted by probe A which is a cDNA fragment corresponding to exon 3 of the murine IFNGR2 gene. The correct targeting was confirmed by blotting the same membrane with a neo cassette probe and was further confirmed by cutting genomic DNA by XbaI and probed by probe B which is a cDNA fragment corresponding to exon 1 of the murine IFNGR2 gene.
B Cell Isolation and Culture.
IgM+ B cells were isolated with a MiniMACS cell sorting column by the standard procedure described by the vendor (Miltenyi Biotec, Auburn, CA). The cells were then cultured at a concentration of 1 × 105 cells/ml and lipopolysaccharide (LPS) added at 10 μg/ml, IL-4 at 50 ng/ml, and IFN-γ at 30 units/ml. Four days later cells were split one to two, and Ig isotypes in the media were measured 2 days later by ELISA.
In Vitro Differentiation of T Cells.
The isolation of naive CD4+ T cells was described previously (38). Briefly, the splenocytes were incubated with anti-CD8, anti-B220, and anti-Mac-1 antibodies. The CD4+ T cells were then enriched by negative depletion of CD8+ T cells, macrophages, and B cells by magnetic beads coated with anti-mouse antibodies (Dynal, Lake Success, NY). The Mel14+ and CD4+ T cells were purified by cell sorting with a flow cytometer (FACStar, Becton Dickinson). Cells (3 × 105 cells/well) were cultured in anti-CD3 antibody- (PharMingen) coated 24-well plates. Various cytokines and antibodies were added to these cultures: murine IL-2 at 1 ng/ml, murine IL-4 at 10 ng/ml, murine IFN-γ (Genzyme) at 10 ng/ml, anti-IL-4 antibodies at 10 μg/ml, and anti-IFN-γ antibody at 15 μg/ml. The cells were cultured for 7 days. Then the cells were plated on a new α-CD3-coated 24-well plate at 5 × 105/well for 2 days.
Protein Immunoprecipitation and Electrophoretic Mobility Shift Assay.
Extracts were precleared by the addition of 3 μl of preimmune serum with protein A–Sepharose (Oncogene Science) beads per 2 mg of whole-cell extract. After mixing at 4°C for 2 h, supernatants were collected and divided into 2-mg aliquots. Antiserum (1:100 dilution; Upstate Biotechnology, Lake Placid, NY) was added to each of these samples and allowed to incubate at 4°C overnight. Subsequently, 15 μl of protein A–Sepharose was added, and samples were agitated for an additional 90 min. The immunoprecipitates were then collected and washed three times in 500 μl of lysis buffer (39). Electrophoretic mobility shift assay was previously described (39).
Northern Blot Analysis.
Northern Blot analysis was carried out according to a standard protocol (40). The DNA fragments used as probes for IRF-1 and GBP-1 expression were amplified by reverse transcriptase–PCR.
Contact Hypersensitivity.
Female IFN-γR2 +/+ and IFN-γR2 −/− mice were sensitized by the epicutaneous application of 400 μl of 0.5% fluorescein isothiocyanate (FITC) to the shaved abdomen. Six days after sensitization, the baseline ear thickness of the mouse’s right ear was measured with a thickness gauge (Meegan Tool Sales, Glastonbury, CT). Following ear baseline measurements, the right ear was epicutaneously treated with 10 μl of 0.5% FITC and the right ear thickness was measured 24 hr later.
RESULTS
Generation of the IFN-γR2 Chain-Deficient Mice.
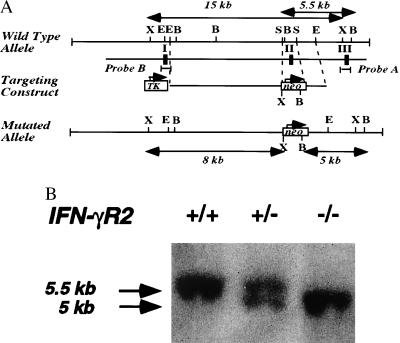
The IFN-γR2-targeting vector (Fig. 1A) was constructed from a 11-kb DNA fragment derived from a mouse 129/Sv genomic library. Within the construct, the second coding exon, which encoded a large region of the extracellular domain of the IFN-γR2 (36), was replaced by a neomycin resistance gene (neo). This DNA construct was introduced into W9.5 ES cells (37) by electroporation, and transfectants were selected with G418 and FIAU. Resistant colonies were screened by Southern blot hybridization with a probe generated from exon 3 (Fig. 1B). Two different clones that had targeted the IFN-γR2 gene by homologous recombination were injected into C57BL/6 blastocysts to generate chimeric mice. Chimeric mice derived from both clones transmitted the mutation to their offspring.
Figure 1.
Gene-targeted mutagenesis of IFNGR2 gene. (A) Schematic drawing of the IFNGR2 gene genomic locus. The abbreviation for restriction enzymes are X: XbaI, E: EcoRI, B: BamHI, S: SmaI. TK is the cassette of herpes thymidine kinase driven by PGK promoter. Probe A was generated by PCR with primers flanking exon 3 of the IFNGR2 gene. Probe B is the PCR-amplified fragment which represents exon 1 of the IFNGR1 gene. (B) The detection of the targeted IFNGR2 gene genomic locus by Southern blot analysis. Genomic DNA generated from mouse tails were digested with BamHI. The Southern blot was hybridized to probe A described above.
In a specific pathogen free facility, mating heterozygotic mice produced offspring with a normal Mendelian segregation pattern. The mutant mice developed normally and all tissues and organs appeared to be intact. Fluorescence-activated cell sorter analysis of thymus, spleen, bone marrow, and peritoneal cells using surface marker such as CD3, CD4, CD8, B220, IgM, CD23, and CD40 did not detect significant changes of major leukocyte populations (data not shown).
Mutation of IFN-γR2 Results in Defective IFN-γ Signaling.
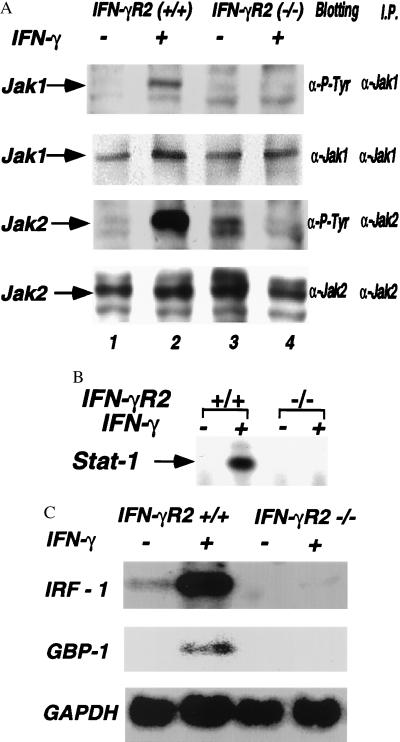
The requirement for Jak1 and Jak2 in IFN-γ signaling has led to a model which proposes that oligomerization of the IFN-γR1 chain (which binds Jak1) and IFN-γR2 chain (which binds Jak2) allows these two kinases to activate each other, thereby initiating a chain of phosphorylation reaction leading to the activation of the Stat1 transcription factor. To determine the effect of the lack of this chain on JAK activation, embryonic fibroblasts from either wild-type or IFN-γR2 −/− mice were cultured with IFN-γ for 15 min. Extracts from these cells were examined for the activation of Jak1 and Jak2. Culture of wild-type fibroblasts with IFN-γ stimulated the phosphorylation of Jak1 and Jak2 (Fig. 2A, lanes 1 and 2). In contrast, −/− cells did not phosphorylate Jak1 or Jak2 in response to IFN-γ (Fig. 2A, lanes 3 and 4). These results demonstrate that IFN-γR2 is required for the activation of both Jak1 and Jak2 in response to IFN-γ. To determine whether the lack of IFN-γR2 also alters activation of the Stat1 transcription factor, splenocytes were isolated from both +/+ and −/− mice and cultured with IFN-γ for 15 min. Extracts isolated from these cells were assayed for activation of Stat1 by electrophoretic mobility shift assay using a probe from the IFN-γ-activating site of the IRF-1 promoter (17). When cells from +/+ mice were cultured with IFN-γ, a Stat1 DNA-binding complex was formed (Fig. 2B, lanes 1 and 2). In contrast, IFN-γ did not activate Stat1 DNA in the −/− splenocytes (Fig. 2B, lanes 3 and 4). These results demonstrate that IFN-γR2 is essential for the IFN-γ induction of both JAK and STAT activation.
Figure 2.
The IFN-γR2 chain disruption results in defective JAK-STAT activation by IFN-γ. (A) IFN-γR2 is required for activation of Jak1 and Jak2 by IFN-γ. Embryonic fibroblasts from both wild-type mice (lanes 1 and 2) and IFN-γR2 −/− mice (lanes 3 and 4) were cultured with IFN-γ (lanes 2 and 4) for 10 min. The extracts from these cells were immunoprecipitated with anti-Jak1 or anti-Jak2 antisera and blotted with either the 4G10 antibody and anti-Jak1 or anti-Jak2 antisera. (B) IFN-γR2 is required for Stat1 activation by IFN-γ. Splenocytes from wild-type mice (lanes 1 and 2) and IFN-γR2 −/− mice (lanes 3 and 4) were either cultured without (lanes 1 and 3) or with IFN-γ (lanes 2 and 4) for 15 min. (C) Total splenocytes were isolated from mice and cultured with IFN-γ for 2 hr. The total RNA was analyzed by Northern blotting with specific IRF-1, GBP-1, and glyceraldehyde-3-phosphate dehydrogenase probes.
Many of the biologic effects induced by IFN-γ require the activation of gene expression. To determine whether the lack of IFN-γR2 alters the induction of gene expression by IFN-γ, splenocytes from these mice were cultured with IFN-γ for 2 hr and the levels of mRNA encoding several genes were examined by Northern blotting. The IRF-1 gene was expressed constitutively in splenocytes isolated from +/+ mice and the level of this mRNA was increased upon culturing with IFN-γ (Fig. 2C, lanes 1 and 2). In contrast, IRF-1 was not constitutively expressed in −/− splenocytes (Fig. 2C, lanes 3) nor was it expressed following incubation with IFN-γ (Fig. 2C, lanes 4). Similarly, IFN-γ induced expression of GBP-1 in splenocytes from +/+ mice, but not in −/− splenocytes. These studies demonstrate that splenocytes from the −/− mice are defective in IFN-γ-induced gene expression.
IFN-γR2 Is Required for the Regulation of Ig Class Switching by IFN-γ.
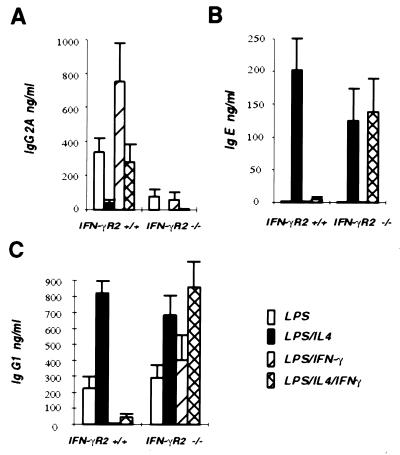
IFN-γ has been shown to induce Ig heavy chain class switching to IgG2a, while it also inhibits class switching to IgE and IgG1 induced by IL-4 (reviewed in ref. 41). To determine whether IFN-γR2 is required for the IFN-γ regulation of class switching, IgM+ B cells were isolated from IFN-γR2 −/− and +/+ mice and were cultured with LPS and different combination of IL-4 and IFN-γ for 6 days. The supernatants from these cultures were harvested and assayed for the concentration of IgG2a, IgG1, and IgE by ELISA (Fig. 3). B cells from +/+ mice cultured with LPS alone produced low levels of IgG2a. When these cells were cultured with LPS plus IFN-γ, the IgG2a secretion was greatly increased. In contrast, the levels of IgG2a produced by B cells isolated from −/− mice cultured under the same conditions were much lower. These B cells produced much less IgG2a when cultured with LPS alone, and, strikingly, IFN-γ did not induce IgG2a production in the LPS cultures of these −/− B cells (Fig. 3A). Therefore, IFN-γR2 was required for effective class switching to IgG2a. Examination of the IgE and IgG1 levels in these cultures also demonstrated that IFN-γR2 was required for the IFN-γ inhibition of IL-4-induced class switching. Thus, although IFN-γ reduced the production of IgE and IgG1 induced by IL-4 in B cells from +/+ mice, it did not reduce the production of IgE or IgG1 induced by IL-4 in B cells isolated from −/− mice (Fig. 3 B and C). These results demonstrated the essential role of IFN-γR2 in IFN-γ-regulated heavy chain class switching in murine B cells.
Figure 3.
Defect in class switching in B cells isolated from IFN-γR2 −/− mice. IgM+ B cells were isolated from total splenocytes and then cultured for 6 days in either LPS, or LPS plus IL-4, or LPS plus IFN-γ, or LPS plus IL-4 plus IFN-γ. The culture supernatants were then collected and assayed for IgG2a (A), IgE (B), and IgG1 (C) production by ELISA.
Disruption of IFN-γR2 Results in Altered Th1-Type Responses.
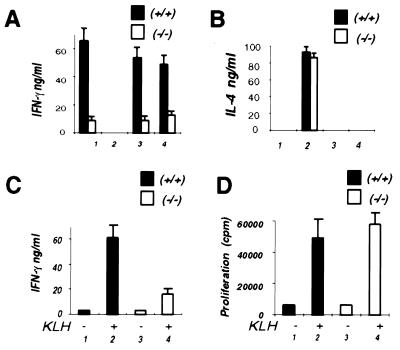
Previous studies have shown that IFN-γR2 is expressed in Th2 but not in Th1 murine T helper cells. To determine whether IFN-γR2 is required for Th1 cell differentiation, naive T helper cells (Mel14+ CD4+) were isolated from both IFN-γR2 +/+ and IFN-γR2 −/− mice and cultured with IL-2 in wells coated with anti-CD3 antibodies (38). To polarize the Th cell populations in these cultures, different cytokines and antibodies to cytokines were added at the initiation of these cultures. Seven days later, these cells were restimulated for 2 days and the cytokine production was analyzed by ELISA. When IL-4 and anti-IFN-γ antibody were added to these cultures, T cells from both mice produced large and similar amounts of IL-4 but no IFN-γ on restimulation (Fig. 4 A and B), demonstrating that differentiation toward a Th2 cell phenotype was not affected by the absence of the IFN-γR2. When naive CD4+ T cells from +/+ mice were differentiated under neutral condition (i.e., in the absence of IL-4, IFN-γ, and anticytokine antibodies), the cells secreted no detectable IL-4 but large quantities of IFN-γ on restimulation (Fig. 4 A and B, condition 1). This was consistent with previous work that T cells cultured under this condition differentiate predominantly toward a Th1 phenotype (38). However, when naive CD4+ T cells from −/− mice were cultured under the same condition, they secreted much less IFN-γ on restimulation (Fig. 4 A and B, condition 1). No IL-4 was produced when T cells from either the wild-type or −/− mice were cultured under this condition. This suggested that under this “default” condition, T cells from −/− mice had a decreased proclivity to differentiate to Th1 cells but did not differentiate to the Th2 phenotype. To further examine Th1 cell differentiation in these mice, naive T cells were cultured with antibodies against IL-4 in the presence of either IFN-γ or IL-12. When naive CD4+ T cells from +/+ mice were cultured with IFN-γ and anti-IL-4 antibodies or IL-12 and anti-IL-4 antibodies, large amounts of IFN-γ were produced (Fig. 4A, conditions 3 and 4). In contrast, when naive CD4+ T cells from −/− mice were cultured under such conditions, T cells secreted much less IFN-γ on restimulation (Fig. 4A, conditions 3 and 4). These results suggest that IFN-γR2 is important for both IFN-γ- and IL-12-induced Th1 differentiation.
Figure 4.
Altered Th1 responses in IFN-γR2 −/− mice. +/+, wild-type mice. −/−, IFN-γR2-deficient mice. (A) IFN-γ production of in vitro differentiated CD4 T cells. Naive (Mel14+) CD4+ T cells were cultured for 7 days in anti-CD3 antibody-coated wells with IL-2 (bar set 1), or IL-2, IL-4, and anti-IFN-γ antibody (bar set 2); or IL-2, IFN-γ, and anti-IL-4 antibody (bar set 3); or IL-12 and anti-IL-4 antibody (bar set 4). The results represent the average of three independent experiments. (B) IL-4 production of the in vitro differentiated CD4+ T cell cultures described above in A. The results represent the average of three independent experiments. (C and D) Antigen-specific T cell cytokine production in local LNCs isolated from KLH-immunized mice. Both wild-type mice (bars 1 and 2) and IFN-γR2 −/− mice (bars 3 and 4) were immunized with KLH in complete Freund’s adjuvant. The LNCs were isolated on day 10 and cultured with (bars 2 and 4) or without KLH (at 100 μg/ml) for 2 days. The supernatants were then assayed for IFN-γ production (C). The duplicated wells of cells were pulsed with 3H on day 2 for 8 hr and proliferation was assayed on day 3 (D). The results represent the average of four independent experiments.
To determine whether IFN-γR2 is required for antigen-specific T cell cytokine production, −/− mice and their wild-type littermates were immunized with the protein antigen keyhole limpet hemocyanin (KLH) in complete Freund’s adjuvant. Ten days after immunization, local lymph node cells (LNCs) were isolated and cultured either with or without KLH for 2 days, and cytokine production was analyzed. LNCs from immunized +/+ mice produced significant levels of IFN-γ when restimulated in vitro with KLH (Fig. 4C, bars 1 and 2). In contrast, the production of IFN-γ in response to KLH was 60% lower in the LNCs isolated from the −/− mice (Fig. 4C, bars 3 and 4). Neither culture produced detectable IL-4. To further explore the immune response in these mice, we examined the ability of these LNCs to proliferate in response to the KLH antigen. Both the LNCs from +/+ mice and those from −/− mice appeared to proliferate in response to the antigen equally, suggesting that the T cells from both mice could respond to the antigen (Fig. 4D). These results are consistent with the results obtained from the in vitro culture experiment and suggest that T cells from −/− mice have a defect in generating Th1-type effector cells.
Compromised Cellular Immunity of IFN-γR2 −/− Mice.
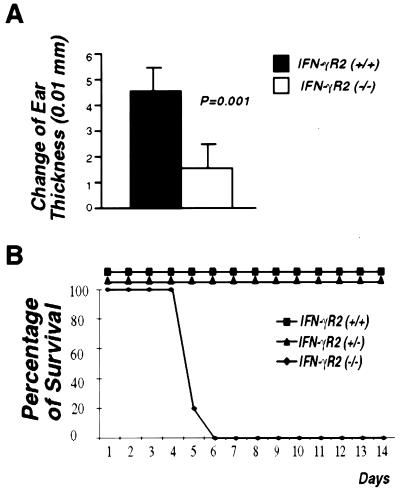
IFN-γ has been shown to promote cell-mediated immune responses (42). To determine whether IFN-γR2 is required for the IFN-γ-mediated cellular immunity in vivo, contact hypersensitivity responses were examined in both wild-type and IFN-γR2 −/− mice. IFN-γR2 −/− mice and their wild-type littermates were sensitized by the epicutaneous application of FITC to the shaved abdomen. Six days after sensitization, contact hypersensitivity was elicited by epicutaneous application of FITC to the ears. As an indication of the contact hypersensitivity response, thickening of ears was measured. In five different experiments, wild-type mice showed strong contact hypersensitivity responses. In contrast, −/− mice had sharply reduced contact hypersensitivity response, with 70% less ear thickening when compared with their wild-type littermates (Fig. 5A).
Figure 5.
Impaired cellular immunity of IFN-γR2 −/− mice. (A) Defective contact hypersensitivity in IFN-γR2 −/− mice. Five 6-wk-old female wild-type mice and five 6-wk-old IFN-γR2 −/− mice were used in this assay. The assay was done as described in Materials and Methods. Contact hypersensitivity is expressed in the change of ear thickness (T). T = (ear thickness 24 hr after elicitation) − (baseline ear thickness). The t test was used in comparison of T in IFN-γR2 +/+ and IFN-γR2 −/− mice. (B) IFN-γR2 −/− mice were susceptible to L. monocytogenes infection. Six-week-old female mice from each genetic background were used. For each genotype, five mice were injected i.p. with 1 × 104 live L. monocytogenes. Percentage of survival represents the percentage of live mice each day.
IFN-γ is important for the activation of the immune response against intracellular parasites such as L. monocytogenes (2). The murine resistance to this microbial pathogen depends on T cell-independent and T cell-dependent mechanisms (43). To determine whether signaling through the IFN-γR2 chain is required for resistance to L. monocytogenes, fifteen 8-wk-old female mice, five from each genomic background, were injected with this pathogen. At the sublethal dose all +/+ and +/− mice survived beyond 14 days, whereas all of the five −/− mice died at day 6 after injection (Fig. 5B). These results demonstrate that the IFN-γR2 chain is required for an effective immune response to L. monocytogenes.
DISCUSSION
In this study, we have established an essential role for the IFN-γR2 chain in IFN-γ function in vivo. In cells isolated from IFN-γR2 −/− mice, Jak2 cannot be activated by IFN-γ, presumably because it is no longer recruited into the receptor complex by the IFN-γR2 chain. In addition, IFN-γ cannot activate Jak1 in these cells although the IFN-γR1 chain, which can bind IFN-γ, is expressed. The requirement for both IFN-γ receptor chains prior to the initiation of signaling is distinct from that for some other cytokine receptor complexes that signal via two distinct receptor chains. For example, homodimerization IL-4 receptor α chain, in the absence of the common γ chain, is sufficient for both the activation of JAK/STAT proteins and the initiation of IL-4-induced gene transcription (44–46). These results suggest that the higher order organization of IL-4 and IFN-γ receptor complexes may be quite distinct.
Disruption of the IFN-γR2 gene also results in a defect in IFN-γ-induced gene expression. This failure to induce transcription is likely due to the inability to activate Stat1 in these cells. It is interesting to note that the constitutive expression of IRF-1 in splenocytes is reduced in the IFN-γR2-deficient mice. Examination of IRF-1-deficient mice has previously demonstrated that these mice contain reduced numbers of CD4−CD8+ αβ T cells and defective NK cells (47, 48). In contrast, our FACS analysis of thymocytes and splenocytes demonstrated that the ratio between CD4+CD8− and CD4−CD8+ αβ T cells in IFN-γR2 −/− mice was similar to that in IFN-γR2 (+/+) mice. These results suggest that IFN-γR2-dependent expression of IRF-1 is not required for the development of CD4−CD8+ and CD4+CD8− αβ T cells.
Disruption of the IFN-γR2 gene also affects the function of B lymphocytes. The ability of B cells to undergo Ig heavy chain class switching is regulated by IFN-γ and IL-4 (41). Furthermore, this regulation of class switching is dependent on the ability of these cytokines to activate germ-line transcription of heavy chain genes (38). IFN-γ promotes IgG2a production in B cells but inhibits switching to IgE and IgG1. The experiments described above demonstrate that the induction of class switching by IFN-γ is dependent on the expression of IFN-γR2. This defect is likely secondary to the inability to activate germ-line transcripts from the IgG2a locus, which is a Stat1-dependent event (P.B.R., unpublished data). In B cells from these mice, IFN-γ also fails to suppress IL-4-induced IgE and IgG1 production. Again, the ability of IFN-γ to suppress IL-4-induced germ-line ɛ and γ1 transcripts is also a Stat1-dependent event (P.B.R., unpublished data). Previously, IFN-γR1-deficient mice have been shown to have a defect in antigen-specific class switching to IgG2a, which could be attributed to a defect in either B cell or T cell compartments (49). Our study demonstrates that B cells from IFN-γR2-deficient mice are unable to class switch to IgG2a in response to IFN-γ.
One of the important findings in these mice is that disruption of the IFN-γR2 −/− gene alters Th1 cell differentiation. This was demonstrated both in vitro by the differentiation of naive CD4+ T cells and in vivo by examining antigen-specific T cell responses. Naive CD4+ T cells from wild-type mice could differentiate to Th1-type T cells either when IFN-γ plus the anti-IL-4 antibody were added to the culture, or when IL-12 and the anti-IL-4 antibody were added to the culture. In contrast, the production of IFN-γ in these cultures was greatly reduced when the naive CD4+ T cells were isolated from the IFN-γR2 −/− mice. The failure of IL-12 to induce potent Th1 differentiation in these cultures may be due to the inability of IFN-γ to maintain IL-12 responsiveness in T cells from IFN-γR2 −/− mice (50). In fact, the level of IL-12Rβ2 in activated CD4+ T cells from IFN-γR2 −/− mice is lower than that of wild-type CD4+ T cells (B.L., unpublished data). This is consistent with a role for IFN-γ in the regulation of IL-12Rβ2 expression (49). Whether decreased levels of IL-12Rβ2 are solely responsible for the defect in Th1 cell polarization is not clear.
Disruption of the IFN-γR2 gene results in a severe defect in cellular immunity in mice as illustrated by assays such as contact hypersensitivity and L. monocytogenes infection. There are several steps in these immune responses that may be defective in these mice. First, macrophage activation induced by IFN-γ is likely lacking in these mice. Another possibility is a defect in NK cell responses to the antigen challenge. Previous studies on IRF-1-deficient mice demonstrate defects in NK cell development. IRF-1 expression is reduced in the IFN-γR2 −/− mice. Therefore, NK cells could potentially be affected in these mutant mice. Third, the ability of T helper cells to differentiate to Th1-type cells is reduced in the IFN-γR2 −/− mice. It is possible that all of these defects contribute to the immunodeficiency in these mice.
The immune deficiencies found in IFN-γ-deficient mice, IFN-γR1-deficient mice, IFN-γR2-deficient mice, Stat1-deficient mice, and humans with a mutation in the IFN-γR1 gene have many similarities (29, 49, 51, 52). All of these defects alter the host resistance to intracellular parasites. Both humans with an early termination signal in the IFN-γR1 gene and IFN-γ-deficient mice are susceptible to mycobacterium infection, whereas Stat1-deficient mice, IFN-γR1-deficient mice, and IFN-γR2-deficient mice are susceptible to L. monocytogenes infection.
Mice with deficiencies in the various components of the IFN-γ signaling pathway appear to differ in their ability to generate a Th1 response. IFN-γR1-deficient mice, although susceptible to Leishmania major infection, are able to generate Th1 cells (53). IFN-γ-deficient mice, on the other hand, fail to generate Th1 cells and mount a Th2 response during L. major infection (54). Interestingly, CD4+ T cells from transgenic mice which express a dominant negative IFN-γR1 gene could develop Th1 cells normally in the presence of IL-12 in an in vitro culture system (55). This is different from the findings in the present study which demonstrate that cultured naive CD4+ T cells isolated from IFN-γR2-deficient mice are impaired in Th1 cell polarization even in the presence of IL-12. The development of congenic mouse strains, each lacking one of the components of the IFN-γ signaling pathway (i.e., IFN-γ, IFN-γR1, IFN-γR2, or Stat1) should help to resolve these seemingly conflicting results.
Acknowledgments
We thank Drs. Sai Kiang Lim, Lixing Xu, and Lin Liu for sharing their expertise on embryonic stem cell culture and blastocyst injection. Thanks to Drs. Alan Stall and Dan Ng for help on FACS analysis. Thanks to Drs. Martin Reichel, Yacov Ron, Ned Braunstein, Miera Harris, and Alexandra Pernis for critical reading of this manuscript. This work was supported by National Institutes of Health Grants CA-46465 (to S.P.), AI-36450 (to S.P.), and AI-39675 (to P.B.R.) and also by American Cancer Society Grant IM-783 (to P.B.R.).
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: IFN-γ, interferon γ; NK, natural killer; IL, interleukin; LPS, lipopolysaccharide; FITC, isothiocyanate; FACS, fluorescence-activated cell sorter; KLH, keyhole limpet hemocyanin; LNC, lymph node cell.
References
- 1.Yoshimoto T, Okamura H, Tagawa Y, Iwakura Y, Nakanishi K. Proc Natl Acad Sci USA. 1997;94:3948–3953. doi: 10.1073/pnas.94.8.3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Farrar M A, Schreiber R D. Annu Rev Immunol. 1993;11:571–611. doi: 10.1146/annurev.iy.11.040193.003035. [DOI] [PubMed] [Google Scholar]
- 3.Pestka S, Langer J A, Zoon K C, Samuel C E. Annu Rev Biochem. 1987;56:727–777. doi: 10.1146/annurev.bi.56.070187.003455. [DOI] [PubMed] [Google Scholar]
- 4.Pestka S, Kotenko S V, Muthukumaran G, Izotova L S, Cook J R, Garotta G. Cytokine Growth Factor Rev. 1997;8:189–206. doi: 10.1016/s1359-6101(97)00009-9. [DOI] [PubMed] [Google Scholar]
- 5.Bach E A, Aguet M, Schreier R D. Annu Rev Immunol. 1997;15:563–591. doi: 10.1146/annurev.immunol.15.1.563. [DOI] [PubMed] [Google Scholar]
- 6.Langer J, Garotta G, Pestka S. Biotherapy. 1996;8:163–174. doi: 10.1007/BF01877201. [DOI] [PubMed] [Google Scholar]
- 7.Rashidbaigi A, Langer J A, Jung V, Jones C, Morse H G, Tischfield J A, Trill J J, Kung H F, Pestka S. Proc Natl Acad Sci USA. 1986;83:384–388. doi: 10.1073/pnas.83.2.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jung V, Rashidbaigi A, Jones C, Tischfield J A, Shows T B, Pestka S. Proc Natl Acad Sci USA. 1987;84:4151–4155. doi: 10.1073/pnas.84.12.4151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aguet M, Dembic Z, Merlin G. Cell. 1988;55:273–280. doi: 10.1016/0092-8674(88)90050-5. [DOI] [PubMed] [Google Scholar]
- 10.Hemmi S, Bohni R, Stark G, Di Marco F, Aguet M. Cell. 1994;76:803–810. doi: 10.1016/0092-8674(94)90355-7. [DOI] [PubMed] [Google Scholar]
- 11.Soh J, Donnelly R J, Kotenko S, Mariano T M, Cook J R, Wang N, Emanuel S, Schwartz B, Miki T, Pestka S. Cell. 1994;76:793–802. doi: 10.1016/0092-8674(94)90354-9. [DOI] [PubMed] [Google Scholar]
- 12.Kotenko S V, Izotova L S, Pollack B P, Mariano T M, Donnelly R J, Muthukumaran G, Cook J R, Garotta G, Silvennoinen O, Ihle J N, Pestka S. J Biol Chem. 1995;270:20915–20921. doi: 10.1074/jbc.270.36.20915. [DOI] [PubMed] [Google Scholar]
- 13.Marsters S A, Pennica D, Bach E, Schreiber R D, Ashkenazi A. Proc Natl Acad Sci USA. 1995;92:5401–5405. doi: 10.1073/pnas.92.12.5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walter M R, Windsor W T, Nagabhushan T L, Lundell D J, Lunn C A, Zauodny P J, Narula S K. Nature (London) 1995;376:217–218. doi: 10.1038/376230a0. [DOI] [PubMed] [Google Scholar]
- 15.Schindler C, Darnell J E J. Annu Rev Biochem. 1995;64:621–651. doi: 10.1146/annurev.bi.64.070195.003201. [DOI] [PubMed] [Google Scholar]
- 16.Watling D, Guschin D, Muller M, Silvennoinen O, Witthuhn B, Quelle F W, Rogers N C, Schindler C, Stark G R, Ihle J N, Kerr I M. Nature (London) 1993;366:166–170. doi: 10.1038/366166a0. [DOI] [PubMed] [Google Scholar]
- 17.Shuai K, Ziemiecki A, Wilks A F, Harpur A G, Sadowski H B, Gilman M Z, Darnell J E. Nature (London) 1993;366:580–583. doi: 10.1038/366580a0. [DOI] [PubMed] [Google Scholar]
- 18.Silvennoinen O, Ihle J N, Schlessinger J, Levy D E. Nature (London) 1993;366:583–585. doi: 10.1038/366583a0. [DOI] [PubMed] [Google Scholar]
- 19.Kotenko S V, Izotova L S, Pollack B P, Muthukumaran G, Paukku K, Silvennoinen O, Ihle J N, Pestka S. J Biol Chem. 1996;271:17174–17182. doi: 10.1074/jbc.271.29.17174. [DOI] [PubMed] [Google Scholar]
- 20.Cook J R, Jung V, Schwartz B, Wang P, Pestka S. Proc Natl Acad Sci USA. 1992;89:11317–11321. doi: 10.1073/pnas.89.23.11317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farrar M A, Campbell J D, Schreiber R D. Proc Natl Acad Sci USA. 1992;89:11706–11710. doi: 10.1073/pnas.89.24.11706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greenlund A C, Farrar M A, Viviano B L, Schreiber R D. EMBO J. 1994;13:1591–1600. doi: 10.1002/j.1460-2075.1994.tb06422.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Decker T, Lew D J, Mirkovitch J, Darnell J E J. EMBO J. 1991;10:927–932. doi: 10.1002/j.1460-2075.1991.tb08026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sakatsume M, Igarashi K, Winestock K D, Garotta G, Larner A C, Finbloom D S. J Biol Chem. 1995;270:17528–17534. doi: 10.1074/jbc.270.29.17528. [DOI] [PubMed] [Google Scholar]
- 25.Wen Z, Zhong Z, Darnell J E J. Cell. 1995;82:241–250. doi: 10.1016/0092-8674(95)90311-9. [DOI] [PubMed] [Google Scholar]
- 26.Singh K, Balligand J L, Fischer T A, Smith T W, Kelly R A. J Biol Chem. 1996;271:1111–1117. doi: 10.1074/jbc.271.2.1111. [DOI] [PubMed] [Google Scholar]
- 27.Argetsinger L S, Norstedt G, Billestrup N, White M F, Carter-Su C. J Biol Chem. 1996;271:29415–29421. doi: 10.1074/jbc.271.46.29415. [DOI] [PubMed] [Google Scholar]
- 28.Visnjic D, Batinic D, Banfic H. Blood. 1997;89:81–91. [PubMed] [Google Scholar]
- 29.Meraz M A, White J M, Sheehan K C, Bach E A, Rodig S J, Dighe A S, Kaplan D H, Riley J K, Greenlund A C, Campbell D, Carver-Moore K, DuBois R N, Clark R, Aguet M, Schreiber R D. Cell. 1996;84:431–442. doi: 10.1016/s0092-8674(00)81288-x. [DOI] [PubMed] [Google Scholar]
- 30.Durbin J E, Hackenmiller R, Simon M C, Levy D E. Cell. 1996;84:443–450. doi: 10.1016/s0092-8674(00)81289-1. [DOI] [PubMed] [Google Scholar]
- 31.Pernis A, Gupta S, Gollob K J, Garfein E, Coffman R L, Schindler C, Rothman P. Science. 1995;269:245–247. doi: 10.1126/science.7618088. [DOI] [PubMed] [Google Scholar]
- 32.Sakatsume M, Finbloom D S. J Immunol. 1996;156:4160–4166. [PubMed] [Google Scholar]
- 33.Skrenta H, Peritt D, Cook J R, Garotta G, Trinchieri G, Pestka S. Eur Cytokine Network. 1996;7:622. [Google Scholar]
- 34.Bach E A, Szabo S J, Dighe A S, Ashkenazi A, Aguet M, Murphy K M, Schreiber R D. Science. 1995;270:1215–1218. doi: 10.1126/science.270.5239.1215. [DOI] [PubMed] [Google Scholar]
- 35.Groux H, Sornasse T, Cottrez F, de Vries J E, Coffman R L, Roncarolo M-G, Yssel H. J Immunol. 1997;158:5627–5631. [PubMed] [Google Scholar]
- 36.Ebensperger C, Rhee S, Muthukumaran G, Lembo D, Donnelly R, Pestka S, Dembic Z. Scand J Immunol. 1996;44:599–606. doi: 10.1046/j.1365-3083.1996.d01-353.x. [DOI] [PubMed] [Google Scholar]
- 37.Kontgen F, Stewart C. Guide to Techniques in Mouse Development. Academic; 1993. pp. 878–890. [Google Scholar]
- 38.Gollob K J, Coffman R L. J Immunol. 1994;152:5180–5188. [PubMed] [Google Scholar]
- 39.Lu B, Reichel M, Fisher D A, Smith J F, Rothman P. J Immunol. 1997;159:1255–1264. [PubMed] [Google Scholar]
- 40.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning. New York: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 41.Coffman R L, Lebman D A, Rothman P. Adv Immunol. 1993;54:229–270. doi: 10.1016/s0065-2776(08)60536-2. [DOI] [PubMed] [Google Scholar]
- 42.Seder R A, Paul W E. Annu Rev Immunol. 1994;12:635–673. doi: 10.1146/annurev.iy.12.040194.003223. [DOI] [PubMed] [Google Scholar]
- 43.Rogers H W, Sheehan K C, Brunt L M, Dower S K, Unanue E R, Schreiber R D. Proc Natl Acad Sci USA. 1992;89:1011–1015. doi: 10.1073/pnas.89.3.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lai S Y, Molden J, Liu K D, Puck J M, White M D, Goldsmith M A. EMBO J. 1996;15:4506–4514. [PMC free article] [PubMed] [Google Scholar]
- 45.Reichel M, Nelson B H, Greenberg P D, Rothman P B. J Immunol. 1997;158:5860–5867. [PubMed] [Google Scholar]
- 46.Fujiwara H, Hanissian S H, Tsytsykova A, Geha R S. Proc Natl Acad Sci USA. 1997;94:5866–5871. doi: 10.1073/pnas.94.11.5866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harada H, Kitagawa M, Tanaka N, Yamamoto H, Harada K, Ishihara M, Taniguchi T. Science. 1993;259:971–974. doi: 10.1126/science.8438157. [DOI] [PubMed] [Google Scholar]
- 48.Duncan G S, Mittrucker H W, Kagi D, Matsuyama T, Mak T W. J Exp Med. 1996;184:2043–2048. doi: 10.1084/jem.184.5.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang S, Hendriks W, Althage A, Hemmi S, Bluethmann H, Kamijo R, Vilcek J, Zinkernagel R M, Aguet M. Science. 1993;259:1693–1694. doi: 10.1126/science.8456301. [DOI] [PubMed] [Google Scholar]
- 50.Szabo S J, Dighe A S, Gubler U, Murphy K M. J Exp Med. 1997;185:817–824. doi: 10.1084/jem.185.5.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dalton D K, Pitts-Meek S, Keshav S, Figari I S, Bradley A, Stewart T A. Science. 1993;259:1739–1742. doi: 10.1126/science.8456300. [DOI] [PubMed] [Google Scholar]
- 52.Newport M J, Huxley C M, Huston S, Hawrylowicz C M, Oostra B A, Williamson R, Levin M. N Engl J Med. 1996;335:1941–1949. doi: 10.1056/NEJM199612263352602. [DOI] [PubMed] [Google Scholar]
- 53.Swihart K, Fruth U, Messmer N, Hug K, Behin R, Huang S, Del Giudice G, Aguet M, Louis J A. J Exp Med. 1995;181:961–971. doi: 10.1084/jem.181.3.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Z E, Reiner S L, Zheng S, Dalton D K, Locksley R M. J Exp Med. 1994;179:1367–1371. doi: 10.1084/jem.179.4.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dighe A S, Campbell D, Hsieh C S, Clarke S, Greaves D R, Gordon S, Murphy K M, Schreiber R D. Immunity. 1995;3:657–666. doi: 10.1016/1074-7613(95)90136-1. [DOI] [PubMed] [Google Scholar]