Abstract
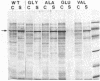
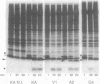
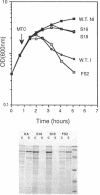
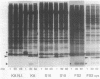
The roles of the various parts of the mature colicin A lysis protein (Cal) in its assembly into the envelope and its function in causing "quasi-lysis," the release of colicin A, and the activation of phospholipase A were investigated. By using cassette mutagenesis, many missense mutations were introduced into the highly conserved portion of the lysis protein. In vitro mutagenesis was also used to introduce stop codons after amino acids 16 and 18 and a frameshift mutation at amino acid 17 of the mature Cal sequence. The processing and modification of the mutants were identical to those of the wild type, except for the truncated Cal proteins, which were neither acylated nor processed. Thus, the carboxy-terminal half of Cal must be present (or replaced by another peptide) for the proper processing and assembly of the protein. However, the specific sequence of this region is not required for the membrane-damaging function of the protein. Furthermore, the sequence specificity for even the conserved amino acids of the amino-terminal half of the protein is apparently exceedingly relaxed, since only those mutant Cal proteins in which a highly conserved amino acid has been replaced by a glutamate were impaired in their function.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachmann B. J. Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol Rev. 1972 Dec;36(4):525–557. doi: 10.1128/br.36.4.525-557.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baty D., Lloubès R., Geli V., Lazdunski C., Howard S. P. Extracellular release of colicin A is non-specific. EMBO J. 1987 Aug;6(8):2463–2468. doi: 10.1002/j.1460-2075.1987.tb02526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernabeu C., Lake J. A. Nascent polypeptide chains emerge from the exit domain of the large ribosomal subunit: immune mapping of the nascent chain. Proc Natl Acad Sci U S A. 1982 May;79(10):3111–3115. doi: 10.1073/pnas.79.10.3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter P., Bedouelle H., Winter G. Improved oligonucleotide site-directed mutagenesis using M13 vectors. Nucleic Acids Res. 1985 Jun 25;13(12):4431–4443. doi: 10.1093/nar/13.12.4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavard D., Baty D., Howard S. P., Verheij H. M., Lazdunski C. Lipoprotein nature of the colicin A lysis protein: effect of amino acid substitutions at the site of modification and processing. J Bacteriol. 1987 May;169(5):2187–2194. doi: 10.1128/jb.169.5.2187-2194.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavard D., Crozel V., Gorvel J. P., Pattus F., Baty D., Lazdunski C. A molecular, genetic and immunological approach to the functioning of colicin A, a pore-forming protein. J Mol Biol. 1986 Feb 5;187(3):449–459. doi: 10.1016/0022-2836(86)90445-6. [DOI] [PubMed] [Google Scholar]
- Cavard D., Lloubès R., Morlon J., Chartier M., Lazdunski C. Lysis protein encoded by plasmid ColA-CA31. Gene sequence and export. Mol Gen Genet. 1985;199(1):95–100. doi: 10.1007/BF00327516. [DOI] [PubMed] [Google Scholar]
- Cole S. T., Saint-Joanis B., Pugsley A. P. Molecular characterisation of the colicin E2 operon and identification of its products. Mol Gen Genet. 1985;198(3):465–472. doi: 10.1007/BF00332940. [DOI] [PubMed] [Google Scholar]
- De Graaf F. K., Oudega B. Production and release of cloacin DF13 and related colicins. Curr Top Microbiol Immunol. 1986;125:183–205. doi: 10.1007/978-3-642-71251-7_11. [DOI] [PubMed] [Google Scholar]
- Ebina Y., Nakazawa A. Cyclic AMP-dependent initiation and rho-dependent termination of colicin E1 gene transcription. J Biol Chem. 1983 Jun 10;258(11):7072–7078. [PubMed] [Google Scholar]
- Hakkaart M. J., Veltkamp E., Nijkamp H. J. Protein H encoded by plasmid Clo DF13 involved in lysis of the bacterial host. I. Localisation of the gene and identification and subcellular localisation of the gene H product. Mol Gen Genet. 1981;183(2):318–325. doi: 10.1007/BF00270635. [DOI] [PubMed] [Google Scholar]
- Jakes K. S., Zinder N. D. Plasmid ColE3 specifies a lysis protein. J Bacteriol. 1984 Feb;157(2):582–590. doi: 10.1128/jb.157.2.582-590.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein P., Somorjai R. L., Lau P. C. Distinctive properties of signal sequences from bacterial lipoproteins. Protein Eng. 1988 Apr;2(1):15–20. doi: 10.1093/protein/2.1.15. [DOI] [PubMed] [Google Scholar]
- Kramer B., Kramer W., Fritz H. J. Different base/base mismatches are corrected with different efficiencies by the methyl-directed DNA mismatch-repair system of E. coli. Cell. 1984 Oct;38(3):879–887. doi: 10.1016/0092-8674(84)90283-6. [DOI] [PubMed] [Google Scholar]
- Lau P. C., Hefford M. A., Klein P. Structural relatedness of lysis proteins from colicinogenic plasmids and icosahedral coliphages. Mol Biol Evol. 1987 Sep;4(5):544–556. doi: 10.1093/oxfordjournals.molbev.a040460. [DOI] [PubMed] [Google Scholar]
- Lloubes R., Baty D., Lazdunski C. The promoters of the genes for colicin production, release and immunity in the ColA plasmid: effects of convergent transcription and Lex A protein. Nucleic Acids Res. 1986 Mar 25;14(6):2621–2636. doi: 10.1093/nar/14.6.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloubès R., Baty D., Lazdunski C. Transcriptional terminators in the caa-cal operon and cai gene. Nucleic Acids Res. 1988 May 11;16(9):3739–3749. doi: 10.1093/nar/16.9.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luirink J., van der Sande C., Tommassen J., Veltkamp E., De Graaf F. K., Oudega B. Effects of divalent cations and of phospholipase A activity on excretion of cloacin DF13 and lysis of host cells. J Gen Microbiol. 1986 Mar;132(3):825–834. doi: 10.1099/00221287-132-3-825. [DOI] [PubMed] [Google Scholar]
- Maget-Dana R., Heitz F., Ptak M., Peypoux F., Guinand M. Bacterial lipopeptides induce ion-conducting pores in planar bilayers. Biochem Biophys Res Commun. 1985 Jun 28;129(3):965–971. doi: 10.1016/0006-291x(85)91985-0. [DOI] [PubMed] [Google Scholar]
- Maget-Dana R., Ptak M., Peypoux F., Michel G. Pore-forming properties of iturin A, a lipopeptide antibiotic. Biochim Biophys Acta. 1985 May 28;815(3):405–409. doi: 10.1016/0005-2736(85)90367-0. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Morlon J., Chartier M., Bidaud M., Lazdunski C. The complete nucleotide sequence of the colicinogenic plasmid ColA. High extent of homology with ColE1. Mol Gen Genet. 1988 Feb;211(2):231–243. doi: 10.1007/BF00330599. [DOI] [PubMed] [Google Scholar]
- Morlon J., Lloubès R., Varenne S., Chartier M., Lazdunski C. Complete nucleotide sequence of the structural gene for colicin A, a gene translated at non-uniform rate. J Mol Biol. 1983 Oct 25;170(2):271–285. doi: 10.1016/s0022-2836(83)80148-x. [DOI] [PubMed] [Google Scholar]
- Pugsley A. P., Cole S. T. An unmodified form of the ColE2 lysis protein, an envelope lipoprotein, retains reduced ability to promote colicin E2 release and lysis of producing cells. J Gen Microbiol. 1987 Sep;133(9):2411–2420. doi: 10.1099/00221287-133-9-2411. [DOI] [PubMed] [Google Scholar]
- Pugsley A. P., Schwartz M. Colicin E2 release: lysis, leakage or secretion? Possible role of a phospholipase. EMBO J. 1984 Oct;3(10):2393–2397. doi: 10.1002/j.1460-2075.1984.tb02145.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugsley A. P., Schwartz M. Expression of a gene in a 400-base-pair fragment of colicin plasmid ColE2-P9 is sufficient to cause host cell lysis. J Bacteriol. 1983 Oct;156(1):109–114. doi: 10.1128/jb.156.1.109-114.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugsley A. P. The immunity and lysis genes of ColN plasmid pCHAP4. Mol Gen Genet. 1988 Feb;211(2):335–341. doi: 10.1007/BF00330613. [DOI] [PubMed] [Google Scholar]
- Sabik J. F., Suit J. L., Luria S. E. cea-kil operon of the ColE1 plasmid. J Bacteriol. 1983 Mar;153(3):1479–1485. doi: 10.1128/jb.153.3.1479-1485.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toba M., Masaki H., Ohta T. Primary structures of the ColE2-P9 and ColE3-CA38 lysis genes. J Biochem. 1986 Feb;99(2):591–596. doi: 10.1093/oxfordjournals.jbchem.a135515. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]