Abstract
The Saccharomyces cerevisiae zinc cluster transcription factors Pdr1 and Pdr3 mediate general drug resistance to many cytotoxic substances also known as pleiotropic drug resistance (PDR). The regulatory mechanisms that activate Pdr1 and Pdr3 in response to the various xenobiotics are poorly understood. In this study, we report that exposure of yeast cells to 2,4-dichlorophenol (DCP), benzyl alcohol, nonionic detergents, and lysophospholipids causes rapid activation of Pdr1 and Pdr3. Furthermore, Pdr1/Pdr3 target genes encoding the ATP-binding cassette proteins Pdr5 and Pdr15 confer resistance against these compounds. Genome-wide transcript analysis of wild-type and pdr1Δ pdr3Δ cells treated with DCP reveals most prominently the activation of the PDR response but also other stress response pathways. Polyoxyethylene-9-laurylether treatment produced a similar profile with regard to activation of Pdr1 and Pdr3, suggesting activation of these by detergents. The Pdr1/Pdr3 response element is sufficient to confer regulation to a reporter gene by these substances in a Pdr1/Pdr3-dependent manner. Our data indicate that compounds with potential membrane-damaging or -perturbing effects might function as an activating signal for Pdr1 and Pdr3, and they suggest a role for their target genes in membrane lipid organization or remodeling.
INTRODUCTION
Evolution equipped the baker's yeast Saccharomyces cerevisiae with a wide range of mechanisms to quickly respond to adverse environmental conditions, including heat, high osmolarity or xenobiotic substances. One mechanism of adapting to toxic compounds is the expression of ATP-binding cassette (ABC) transporters, which remove cytotoxic substances from the cytoplasm either to the vacuole or the extracellular space (Wolfger et al., 2001; Sipos and Kuchler, 2006). The homologous Zn(II)2Cys6 zinc cluster transcription factors Pdr1 and Pdr3 are major regulators of many ABC transporters. Zn(II)2Cys6 zinc cluster transcription factors are a class of proteins strictly specific for fungi (MacPherson et al., 2006). They share a characteristic domain structure and mostly recognize CGG triplets as dimers. Pdr1 and Pdr3 form homo- and heterodimers, which recognize pleiotropic drug resistance elements (PDREs) (Mamnun et al., 2002). Studies of genome-wide mRNA profiles of constitutively active Pdr1 and Pdr3 alleles and analysis of the transcriptional response to certain drugs and toxins explored their target gene spectrum (DeRisi et al., 2000; Devaux et al., 2001, 2002). These include ABC transporters PDR5 (Decottignies et al., 1994; Katzmann et al., 1994), PDR10, PDR15 (Wolfger et al., 1997), SNQ2 (Mahé et al., 1996b), YOR1 (Hallström and Moye-Rowley, 1998) and other genes involved in detoxification. The specific function of Pdr1 and Pdr3 is not entirely redundant, because loss of mitochondrial function causes high expression of Pdr5 involving only Pdr3 (Hallström and Moye-Rowley, 2000; Devaux et al., 2002).
Pdr1 is active under normal growth conditions, because its target gene PDR5 is highly expressed, suggesting a partly constitutive role (Mamnun et al., 2004). Although Pdr1 and Pdr3 gain-of-function mutants show higher PDR gene expression and corresponding drug resistance phenotypes, a dynamic regulation in response to drugs remained an open question. First, indications for such a response came from reports showing rapid induction of certain PDR genes in response to treatment with the herbicide by 2,4-dichlorophenoxy acetic acid (2,4-D) (Teixeira and Sa-Correia, 2002; Teixeira et al., 2004, 2006) and the drugs benomyl and fluphenazine (Fardeau et al., 2007). Pdr1 and Pdr3 activity might, therefore, be regulated directly by small molecules, similar to other fungal zinc cluster transcription factors such as Gal4 (Yano and Fukasawa, 1997), Leu3 (Guo and Kohlhaw, 1996; Wang et al., 1999), Put3 (Des Etages et al., 2001), and Ppr1 (Flynn and Reece, 1999, Sellick and Reece, 2005; Reece et al., 2006). However, for Pdr1 and Pdr3, substances identified as activators in previous studies are structurally and functionally unrelated. Hence, a general chemical property or a common biochemical response shared by them might trigger activation.
In this study, we investigated the global transcriptional response to 2,4-dichlorophenol (DCP) and the nonionic detergent polyoxyethylene-9-laurylether (POELE). These and other membrane-perturbing agents such as lysophospholipids and benzyl alcohol strongly and rapidly induce the expression of the Pdr1 and Pdr3 target genes. The activation of a PDRE-regulated reporter gene suggests the direct activation of Pdr1 and Pdr3. We further show that their targets Pdr5 and Pdr15 are highly induced by various membrane-damaging agents and confer resistance to these compounds. Our data suggest that hydrophobic substances might constitute generic Pdr1/Pdr3-activating signals and that a possible physiological role for their targets Pdr15 or Pdr5 could be in membrane lipid organization or lipid bilayer remodeling after membrane disturbances.
MATERIALS AND METHODS
Yeast Strains, Plasmids, Growth Conditions, and Cytotoxicity Assays
Rich medium (YPD) and synthetic medium (SC), supplemented with appropriate auxotrophic components, were prepared as described previously (Kaiser et al., 1994). Unless otherwise indicated, all yeast strains were grown routinely at 30°C. Tests for drug resistance phenotypes were performed with cells grown to the exponential growth phase and diluted to an OD600 of 0.2. Identical volumes of cultures and 1:10 and 1:100 serial dilutions were spotted onto agar plates containing the indicated concentrations of drugs. The compounds DCP, POELE, and lysophosphatidylcholin (lysoPC) were purchased from Sigma-Aldrich (St. Louis, MO).
Yeast strains and plasmids used in this study are listed in Table 1. Strain YYA100 (pdr1Δ::KanMx6, pdr3Δ::His3Mx6) was obtained by genomic integration of appropriate polymerase chain reaction (PCR) products synthesized from the plasmids pFA6a-HIS3Mx6 and pFA6a-KanMx6 (Longtine et al., 1998). For YHW4 and YHW5, W303-1A and W303-1A msn2Δ msn4Δ cells were transformed with a PCR fragment synthesized from plasmid pFA6a-3HAKanMx6. Correct genomic integration of all fragments was verified by PCR. Plasmid pHWZ15z was generated by cloning the 836bp SmaI–EcoRV fragment from the PDR15 promoter into the SmaI site of YEp368 as described previously (Wolfger et al., 1997).
Table 1.
Yeast strains and plasmids used in this study
| Yeast strain | Relevant genotype | Source |
|---|---|---|
| W303-1Aa | MATaura3-1 leu2-3,112 his3-11,15 trp1-1 ade2-1 can1-100 | |
| NRY201a | pdr5Δ::HIS3 | This study |
| NRY212a | pdr15Δ::TRP1 | This study |
| NRY227a | pdr5Δ::HIS3 pdr15Δ::TRP1 | This study |
| YYA100a | pdr1Δ::Kan-Mx6 pdr3Δ::HIS3-Mx6 | This study |
| W303 msn2Δmsn4Δa | msn2Δ::HIS3 msn4Δ::TRP1 | Görner et al. (1998) |
| YHW4a | PDR15-3HA::Kan-Mx6 | This study |
| YHW5a | msn2Δ::HIS3 msn4Δ::TRP1 PDR15-3HA::Kan-Mx6 | This study |
| FY1679-28cb | MATaura3-52 his3-Δ200 leu2-Δ1 trp1-Δ63 | |
| naΔ1b | pdr1Δ::TRP1 | Delaveau et al. (1994) |
| naΔ3b | pdr3Δ::HIS3 | Delaveau et al. (1994) |
| naΔ1Δ3b | pdr1Δ::TRP1 pdr3Δ::HIS3 | Delaveau et al. (1994) |
| Plasmid | Relevant inserts |
|
| pDK52 | CYC1 promoter, 3xPDRE | Katzmann et al. (1996) |
| pDK53 | CYC1 promoter, 3xmPDRE | Katzmann et al. (1996) |
| pHW-15z | PDR15 promoter-LacZ | This study |
| pADH1-Msn2-GFP | ADH1 promoter-Msn2-GFP | Görner et al. (1998) |
a,b Isogenic set of two parent yeast strains.
Preparation of Extracts and Immunoblotting
For immunoblot analysis, 5 OD600 equivalents of cells were harvested, diluted in 1 ml of water, and incubated on ice with 150 μl of YEX lysis buffer (1.85 M NaOH and 7.5% β-mercaptoethanol) for 10 min. Proteins were precipitated by adding 150 μl of 50% trichloroacetic acid for 10 min. Samples were centrifuged at 13,000 × g, and the pellet was resuspended in sample buffer (40 mM Tris-HCl, pH 6.8, 8 M urea, 5% SDS, 0.1 mM EDTA, 0.1 g/l bromphenol blue, and 1% β-mercaptoethanol). Proteins were separated by SDS-polyacrylamide gel electrophoresis and blotted on nitrocellulose membranes. Pdr5 was immunodetected using the polyclonal anti-Pdr5 antibodies serum Gaston 6 (Egner et al., 1995) and Pdr15-hemagglutinin (HA) with monoclonal anti-HA antibody 16B12 (BAbCO, Richmond, CA).
β-Galactosidase Assays and Fluorescence Microscopy
β-Galactosidase measurements were carried out as described previously (Kaiser et al., 1994). Pdr5-green fluorescent protein (GFP) fluorescence studies were done in living cells grown on YPGE for 6 h, centrifuged, and shifted back to YPD medium. Msn2-GFP fluorescence analysis was done in exponentially growing cells. All media were supplemented with adenine. GFP was visualized using a Zeiss Axioscope II microscope (Carl Zeiss, Jena, Germany). Images were obtained using a Quantix charge-coupled device camera (Roper Scientific, Trenton, NJ) with IPLab software (Spectra Services, Webster, NY).
Northern Blot Analysis
RNA preparation and separation on agarose gels was done exactly as described previously (Wolfger et al., 1997). The membrane was prehybridized in 10 ml of 10× Denhardt's buffer (1 g of Ficoll 400, 1 g of polyvinylpyrrolidone, and 1% [wt/vol] bovine serum albumin fraction V), 2× standard saline citrate (SSC), 1% SDS, and 20 μg/ml salmon sperm DNA for 3–6 h at 65°C. The respective probes were labeled with a MegaPrime labeling kit according to the manufacturer (GE Healthcare, Chalfont St. Giles, United Kingdom) and directly added to the prehybridization solution after purification on a Sephadex G-25 mini column and subsequent heat denaturation. After an overnight incubation at 65°C, membranes were washed three times with 2× SSC, 1% SDS and three times with 1× SSC, 1% SDS at 65°C, and then they were exposed to an x-ray film or analyzed using a PhosphoImager (Storm 1840; GE Healthcare).
DNA Microarray Profiling Experiments
All DNA microarray experiments and data mining were carried out in compliance with the suggested MIAME standards (Brazma et al., 2001). Cells from overnight cultures in YPD were diluted into fresh YPD to an OD600 of 0.1 and grown at 30°C until an OD600 of ∼1 was reached. Cultures were split and 0.3 mM DCP or 0.1 mM POELE was added to one-half of the culture. After 20 min, both untreated and treated cultures were harvested by centrifugation at room temperature (2 min at 4000 × g); cells were quickly washed in ice-cold water, reharvested at 4°C, and frozen at −80°C.
For isolation of total RNA from yeast, frozen pellets equivalent of 10–20 OD600 cells were suspended in 200 μl of cold TES buffer (10 mM Tris-HCl, pH 7.4, 1 mM EDTA, 300 mM NaCl, and 0.2% SDS), and an equal amount of phenol saturated with 10 mM Tris-HCl, pH 6.7, was added. The sample was mixed thoroughly and incubated at 65°C for 1 h, and then it was chilled on ice and centrifuged for 10 min. The aqueous phase was extracted with phenol-chloroform, twice with chloroform, and ethanol precipitated. The pellet was washed with 70% ethanol, dried briefly, dissolved in sterile water at 56°C, and the concentration was determined by spectrophotometry at 260 nm in TE buffer (10 mM Tris-HCl, 1 mM EDTA), pH 7.0.
For cDNA synthesis and labeling, 20 μg of total RNA was transcribed into cDNA by using 200 U of SuperScript II reverse transcriptase (Invitrogen), including either cyanine (Cy)3-dCTP or Cy5-dCTP (GE Healthcare). Labeled cDNAs were pooled; the RNA was removed by hydrolysis in 50 mM NaOH at 65°C for 15 min. The solution was neutralized with acetic acid, and cDNA was precipitated with an equal volume of isopropanol. The pellet was washed briefly in 70% ethanol and finally dissolved in 5 μl of sterile distilled water.
Glass-spotted microarrays (Ontario Cancer Institute, Toronto, ON, Canada; http://www.microarrays.ca/) containing PCR fragments of 6144 predicted S. cerevisiae open reading frames (ORFs) in duplicates were used for the expression profiling. Hybridization was performed in a total volume of 60 μl in DigEasyHyb solution (Roche Diagnostics, Indianapolis, IN) with 0.1 mg/ml salmon sperm DNA (Sigma-Aldrich) as carrier at 37°C for 14–16 h. Microarrays were disassembled in 1× SSC, washed two times in 1× SSC, 0.1% SDS at 50°C for 20 min, followed by a 1-min wash in 1× SSC at room temperature. Slides were spun dry for 5 min at 500 rpm in a tabletop centrifuge at room temperature. Slides were scanned on an Axon4000B scanner (Axon Instruments, Foster City, CA) and analyzed using the GenePix Pro4.1 software (Axon Instruments). Microarrays and all experimental protocols were from the Ontario Cancer Institute (http://www.microarrays.ca/).
Analysis of Microarray Data
The raw data set of this study is available as supplemental material and has been deposited online at arrayexpress (http://www.ebi.ac.uk/arrayexpress/) under the accession number E-MEXP-865. Microarrays were analyzed with GenePixPro4.1 by using standard parameters. For individual microarrays, the intensity of the two fluorescent channels was normalized to the mean of ratio of medians of all unflagged features by using the GenePix Pro4.1 normalization option. Values of not found features were excluded from further analysis. Mean ratios were calculated for features with at least four data points, and their quality was approximated by their coefficient of variation (CV) values excluding values smaller than 1. Genes labeled as dubious ORFs in Saccharomyces Genome Database (SGD; http://www.yeastgenome.org/) were removed from analysis, and several ORFs assigned in SGD recently are not present on the microarrays. The resulting filtered values were normalized by addition of a constant to set the median to the log2 transformed median of ratios values to zero. The normalized values used for further analysis are available as supplementary file. Cluster analysis was performed using the cluster3 and visualized with TreeView (both available at http://rana.stanford.edu/software). Significant associations to either gene ontology (GO)-terms or transcription factors were collected with the T-Profiler (Boorsma et al., 2005). Values of genes associated with the most significant terms were visualized by Cluster analysis by using complete linkage and correlation as similarity metric. The cluster results were also confirmed by K-means clustering. The p values of overlapping genes sets were calculated assuming a hypergeometric distribution. The effect of the absence of the transcription factors Pdr1/pdr3 for DCP gene regulation the ratios of the wild-type versus mutant was calculated, and induced genes with a twofold change were selected. The log2-transformed values were included but not used for cluster analysis by setting their weight to zero. Normalized values with updated canonical gene names are available in plain text format as Supplementary Material as well as the text versions of the Cluster3 output files corresponding to the figures.
Lipid Analysis and Trinitrobenzene Sulfonic Acid (TNBS) Labeling
Cells were grown in 1 ml of YPD supplemented with 50–250 μCi of [32P]phosphate (10 mCi/ml, PerkinElmer-Cetus, Boston, MA) for 15–18 h to permit labeling of phospholipids at constant specific activity (Chang et al., 1998). For reaction of whole yeast cells with TNBS, a previously published procedure (Siegmund et al., 1998) was followed with minor modifications. After labeling, cells were washed (2 × 1 ml) in ice-cold TNBS buffer (120 mM NaHCO3, pH 8.4, and 40 mM NaCl) and resuspended in 1 ml of TNBS buffer. Aliquots of 31 μl of a 5% aqueous solution of TNBS (Sigma-Aldrich) were added, and cells were incubated on ice for 1 h with brief vortex mixing every 20 min. Cells were then washed (3 × 1 ml) in ice-cold TNBS buffer and resuspended in 600 μl of CHCl3:MeOH:0.1N HCl (1:2:0.8) containing carrier lipid (Chang et al., 1998). Cells were lysed by vortexing with glass beads for 3 min, and 200 μl each CHCl3 and HCl/NaCl (0.1 N/0.5 M) were added. The resulting mixture was centrifuged for 2 min to give good phase separation, and the organic phase was removed and analyzed by two-dimensional (2-D) thin layer chromatography (TLC) with dimension I as CHCl3:MeOH:glacial acetic acid (65:25:10) and dimension II as CHCl3:MeOH:88% formic acid (65:25:10) (Esko and Raetz, 1980) by using nonradioactive trinitrophenyl-phosphatidylethanolamine (TNP-PE) as a standard. TLC plates were used to expose phosphor storage screens that were then quantitated using PhosphorImager with ImageQuant software (GE Healthcare). The amount of PE exposed on the outer leaflet of the plasma membrane was calculated as TNP-PE/(TNP-PE + unreacted PE). TNP derivatives of phosphatidylserine were not observed. This assay produced somewhat variable results among different data sets, apparently due to slight differences in the buildup of dead cells with different starter cultures and different aliquots of YPD, but within an individual set the data were quite precise.
TNP-PE was synthesized according to a published procedure (Gordesky et al., 1973) with minor modifications. We dissolved 25 mg of PE in 2.5 ml of CHCl3 and added it to 2.5 ml of MeOH, 1 ml of CHCl3, 250 μl of 5% aqueous NaHCO3, and 390 μl of 5% aqueous TNBS. This mixture was allowed to react at room temperature for 2 h under argon, and it was then transferred to 2 ml of CHCl3 and 20 ml of saturated aqueous NaCl. The organic phase was then acidified with aqueous 1 N HCl by using the color change of unreacted TNBS as an indicator, followed by an additional extraction with 5 ml of carbonate puffer. The organic phase was then dried over Na2SO4 and stored at −20°C under argon, in which condition it was stable for several months as assessed by 2-D TLC.
RESULTS
DCP and Nonionic Detergents Induce PDR and Other Stress Responses
Exposure to 2,4-D, a major constituent of Agent Orange, was suggested to cause a rapid transcriptional response of yeast cells mediated by general stress transcription factors Msn2 and Msn4, the PDR-mediating transcription factors Pdr1 and Pdr3, and the oxidative stress response transcription factor Yap1 (Teixeira and Sa-Correia, 2002; Simoes et al., 2003; Teixeira et al., 2004, 2006). Here, we extend these studies to analyze a possible Pdr1/Pdr3-regulated response. The metabolic fate of 2,4-D in yeast cells is not documented. Enzymatic degradation to glyoxylate and DCP may involve α-ketoglutarate dioxygenase (http://umbbd.msi.umn.edu/) (Ellis et al., 2006). However, such an activity has not yet been described for S. cerevisiae. We, therefore, determined transcript profiles for the related compound DCP. Chemical structures of compounds used in this study are depicted in Supplemental Figure S1. Exponentially growing cells were treated with 0.3 mM DCP for 20 min, because this concentration triggered a strong transcriptional response in previous experiments, causing a transient growth delay for ≈1 h without changing the final growth rate. Total RNA of treated and untreated control was extracted and used as template for synthesis of Cy3- and Cy5-labeled cDNA, which was hybridized to whole genome microarrays. Data sets of more than three independent arrays were analyzed, allowing for the identification of 103 genes induced more than twofold. To investigate the role of Pdr1 and Pdr3, we repeated the experiment by using a pdr1Δ pdr3Δ double deletion strain. Comparison of the obtained results with the 2,4-D data revealed a similar pattern of gene expression to DCP (Supplemental Figure S2). Some 37 out of 161 genes were more than twofold up-regulated by 2,4-D (Teixeira et al., 2006), and 147 genes were more than 1.6-fold up-regulated by DCP (p < 2 × 10−26). Inclusion of the pdr1Δ pdr3Δ data and cluster analysis highlighted the PDR-specific response for both compounds. These data show a rapid Pdr1/Pdr3-dependent transcriptional response similar in extent to other stress responses. A similar pattern of Pdr1/Pdr3-regulated genes has been reported with the drugs benomyl, fluphenazine (Fardeau et al., 2007), and linoleic acid hydroperoxide (Alic et al., 2003). As a trigger of Pdr1/Pdr3 activation, a rather high hydrophobicity or amphipatic nature of compounds stands out as the main common feature of all mentioned substances (Supplemental Figure S1), suggesting a possible membrane perturbance effect of these compounds. For example, DCP might interfere with membrane lipid integrity similar to 2,4-D (Suwalsky et al., 1996; Kaioumova et al., 2001a,b; Tuschl and Schwab, 2003). To test whether membrane damage could be an inducing cue for Pdr1/Pdr3, we used the nonionic detergent POELE for a further set of microarray experiments. The structure of POELE is unrelated to any of the drugs tested so far in connection with Pdr1/Pdr3 (Supplemental Figure S1). Cells in the exponential growth phase (A600 = 1) were treated with 0.1 mM POELE for 20 min, and mRNA profiles were determined by hybridization to microarrays. As for DCP, this concentration was chosen for POELE because of maximal transcriptional response and only slight delay of growth for ≈1 h. Some 193 genes were induced by more than twofold. Raw and filtered data are available as supplemental files as indicated in the figure legends. The entire filtered and normalized data sets for POELE and DCP in wild-type and DCP in pdr1Δ pdr3Δ double deletion strains were analyzed by T-Profiler (http://www.t-profiler.org/; Boorsma et al., 2005) for overrepresented transcription factor binding sites and significant association to GO-terms (Tables 2 and 3). Most significantly represented motifs for DCP included PDR, Msn2/Msn4, Hsf1 (heat-shock element [HSE]), Rpn4, and Ino4. The significant GO-terms are summarized in Table 3, and include “response to stress and abiotic stimulus”, heat shock, and terms associated with proteasome-related degradation. Therefore, the transcription factor motifs and the GO-terms are in good agreement.
Table 2.
T-Profiler analysis for motifs
| Motif | Name | DCP t value | DCP-pdr1/3 t value | POELE t value |
|---|---|---|---|---|
| TCCGYGGA | PDR3 | 11.58 | −0.34 | 5.84 |
| TCCGYGGR | PDR-like | 10.2 | 1.19 | 4.3 |
| TCCGCGG | PDR-like | 5.37 | 1.16 | 3.6 |
| CCCCT | MSN2-4 | 4.47 | −3.21 | 10.26 |
| AGGGG | MSN2-4 | 5.66 | −2.63 | 6.57 |
| TATAWAW | TBP | 5.34 | −0.29 | 5.03 |
| TTCTRGAA | HSF1 | 4.27 | 3.78 | 3.35 |
| GGTGGCRA | RPN4 | 4.02 | 2.26 | 1.48 |
| GCAYGTG | INO4 | 3.8 | 2.78 | −0.42 |
| TAW4TAGM | RLM1 | 3.29 | 3.65 | 0.97 |
| TATGACG | OSMO | 2.51 | −0.37 | 3.59 |
| RMACCCA | RAP1 | −1.29 | 2.09 | −5.58 |
| TTTCGCG | SWI4 | −1.88 | −1.04 | −3.65 |
| TTCTCN6CGC | Novel filament | −3.04 | −3.8 | 1.94 |
| GCGATGAGMTGARAW | rRNA | −4.83 | −0.47 | −2.53 |
| CCRTACA | RAP1 | −5.55 | −2.51 | −2.8 |
| CGATGAG | PAC | −7.85 | −0.25 | −6.09 |
| AAAATTT | rRPE | −10.51 | −2.23 | −9.67 |
Motifs and the name of the transcription factor found to display a significant change in the DCP and POELE early response are indicated, along with the t values associated with each motif (http://www.t-profiler.org/). Input files were POELE-all and DCP-all data available as Supplemental Material.
Table 3.
Selected results from T-Profiler analysis (http://www.t-profiler.org/) for GO-terms associated with POELE and DCP data sets
| T-Profiler analysis | t value | E value | Median | ORF |
|---|---|---|---|---|
| POELE | ||||
| Response to stress | 6.32 | 3.60E-07 | 0.215 | 266 |
| Heat-shock protein activity | 5.46 | 6.60E-05 | 0.901 | 16 |
| Response to stimulus | 5.41 | 8.80E-05 | 0.151 | 358 |
| Oxidoreductase activity, peroxide as acceptor | 4.77 | 2.60E-03 | 0.888 | 13 |
| DCP | ||||
| Oxidoreductase activity, acting on CH-OH groups | 5.78 | 1.00E-05 | 0.316 | 64 |
| Monosaccharide metabolism | 5.4 | 9.30E-05 | 0.289 | 65 |
| Response to stimulus | 5.23 | 2.40E-04 | 0.076 | 391 |
| Response to abiotic stimulus | 5.01 | 7.60E-04 | 0.141 | 168 |
| Response to external stimulus | 4.98 | 8.80E-04 | 0.134 | 179 |
| Aryl-alcohol dehydrogenase activity | 4.93 | 1.10E-03 | 1.061 | 7 |
| Heat-shock protein activity | 4.77 | 2.60E-03 | 0.591 | 16 |
| Aldehyde metabolism | 4.64 | 4.80E-03 | 0.552 | 17 |
| Proteasome complex (sensu Eukarya) | 4.62 | 5.30E-03 | 0.316 | 42 |
| Endopeptidase activity | 4.56 | 7.10E-03 | 0.298 | 45 |
| Response to chemical substance | 4.42 | 1.40E-02 | 0.175 | 97 |
| Protein catabolism | 4.37 | 1.70E-02 | 0.146 | 124 |
| Peptidase activity | 4.36 | 1.80E-02 | 0.178 | 92 |
| Proteasome core complex (sensu Eukarya) | 4.24 | 3.10E-02 | 0.521 | 16 |
| Modification-dependent protein catabolism | 4.18 | 4.00E-02 | 0.173 | 89 |
| Alcohol metabolism | 4.17 | 4.10E-02 | 0.144 | 115 |
| Proteolysis and peptidolysis | 4.17 | 4.10E-02 | 0.148 | 111 |
Input files were POELE-all and DCP-all data sets.
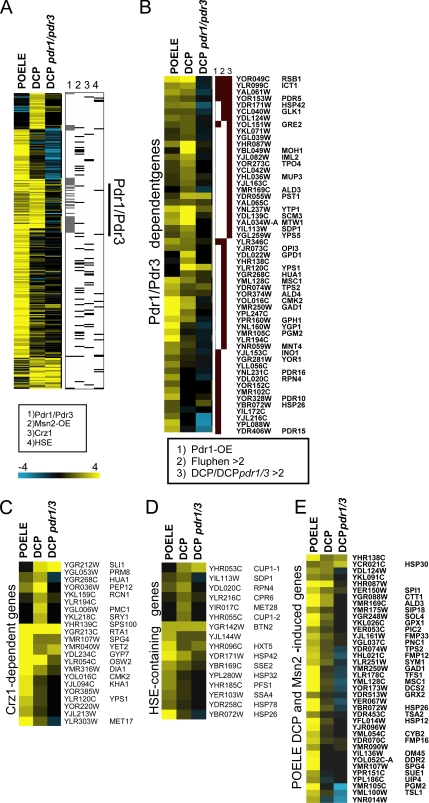
Further analysis was done with a core DCP/POELE gene set selected as more than twofold induced in either condition. Cluster analysis of this core set indicated a large but qualitative distinct overlap between DCP and POELE response (p < 4 × 10−10 for a 2-fold cut-off), further highlighting the Pdr1/Pdr3-dependent subset. Notably, POELE-induced genes were more frequently associated with Msn2/Msn4, Crz1 and contain HSEs (Figure 1A).
Figure 1.
Genome-wide transcriptional response to POELE and DCP treatment. Cells were treated with 0.3 mM DCP or 0.1 mM POELE for 20 min or left untreated. mRNA profiles were determined by hybridization to genome-wide yeast microarrays. (A) Specific and overlapping responses to POELE and DCP: hierarchical clustering of core POELE and DCP gene set selected as more than twofold induced in either POELE- or DCP-treated cells. Genes are indicated with binding sites and/or controlled by other transcription factors such as Pdr1/Pdr3 (Fardeau et al., 2007), Msn2/Msn4 (Chua et al., 2006), Crz1 (Yoshimoto et al., 2002), and Hsf1 (Lee et al., 2002). (B) Pdr1/Pdr3-regulated genes are induced by DCP and POELE. Genes were selected by twofold induction DCP with >1.5-fold difference to the pdr1Δ pdr3Δ deletion mutant values. Reported Pdr1/Pdr3 target genes were selected according to their induction by Pdr1 overexpression (Devaux et al., 2001) and by fluphenazine (Fardeau et al., 2007). DCP-induced genes dependent on Pdr1/Pdr3 include known PDR-target genes, as well as those induced by fluphenozine or Pdr1 overexpression. (C) Some 22 of 98 (Yoshimoto et al., 2002) Crz1-dependent genes are also induced by POELE treatment and weaker by DCP. DCP induction is not changed in wild-type versus Pdr1/Pdr3 deletion mutant. (D) HSE-containing genes in the POELE data set as identified by T-Profiler show in both POELE- and DCP-enhanced expression but no Pdr1/Pdr3 dependence is noted. (E) POELE/DCP-regulated genes also targeted by Msn2/Msn4 were selected from the Msn2/Msn4 overexpression data (Chua et al., 2006). Of 72 genes induced >1.5-fold by Msn2/Msn4 overexpression, 40 are present among the 193 core POELE/DCP-induced set (>2). The values of these genes treated with DCP are generally lower but also significantly increased according to T-Profiler. Files with the normalized values used to generate the figures are available as supplemental material and as Custer3 result files.
The subset of Pdr1/Pdr3-target genes was deduced from the lack of induction by DCP in the pdr1Δ pdr3Δ double deletion strain. This set included and extended those genes identified by Pdr1 overexpression (Devaux et al., 2001) or by fluphenazine treatment (Fardeau et al., 2007) (Figure 1B). Several Crz1 target genes were identified in the benomyl and fluphenazine response (Fardeau et al., 2007). Out of 98 reported Crz1-regulated genes (Yoshimoto et al., 2002), some 22 were also present in the core set (p < 2 × 10−6; Figure 1C).
T-Profiler analysis reported a significant presence of Msn2/Msn4 binding sites in genes induced by DCP, POELE, and 2,4-D treatment (Table 2 and Supplemental Figure S2). This is in line with the activation of Msn2/Msn4 by membrane-perturbing agents such as 2,4-D as demonstrated previously (Moskvina et al., 1999). Motifs presumably recognized by Msn2/Msn4 are either present in too many genes (CCCCT) or in to few (MAGGGGSGG). We used the Msn2 and Msn4 overexpression data sets as a reference for identifying Msn2/Msn4-regulated genes (Chua et al., 2006). Interestingly, induction by DCP of nearly all Msn2/Msn4 target genes was reduced in the pdr1Δ pdr3Δ double deletion strain (Figure 1E). A similar effect was not observed for Crz1, Hsf1, Ino4, and Rpn4, and their target genes were not affected by the lack of Pdr1/Pdr3 (Figure 1, C and D, and Supplemental Figure S2, B–D). The reason for this observation and apparent cross talk between Pdr1/Pdr3 and Msn2/Msn4 is unclear but might be an indirect effect of the pdr1Δ pdr3Δ mutation. As observed in many other acute stress conditions (Gasch et al., 2000), POELE and DCP treatment repressed many ribosomal protein genes more than twofold. This was also detected by T-Profiler analysis (Tables 2 and 3).
Together, exposure of cells to DCP or the nonionic detergent POELE resulted in activation of genes whose expression is mainly regulated through Pdr1/Pdr3 and Msn2/Msn4, but also included Rpn4, Crz1, and Hsf1 target genes. Furthermore, we demonstrate that not only drugs but also detergents activate Pdr1/Pdr3, implying a function of PDR-target genes such as ABC transporters in controlling membrane lipid homeostasis.
Pdr1 and Pdr3 Are Required for Resistance to Membrane-perturbing Agents
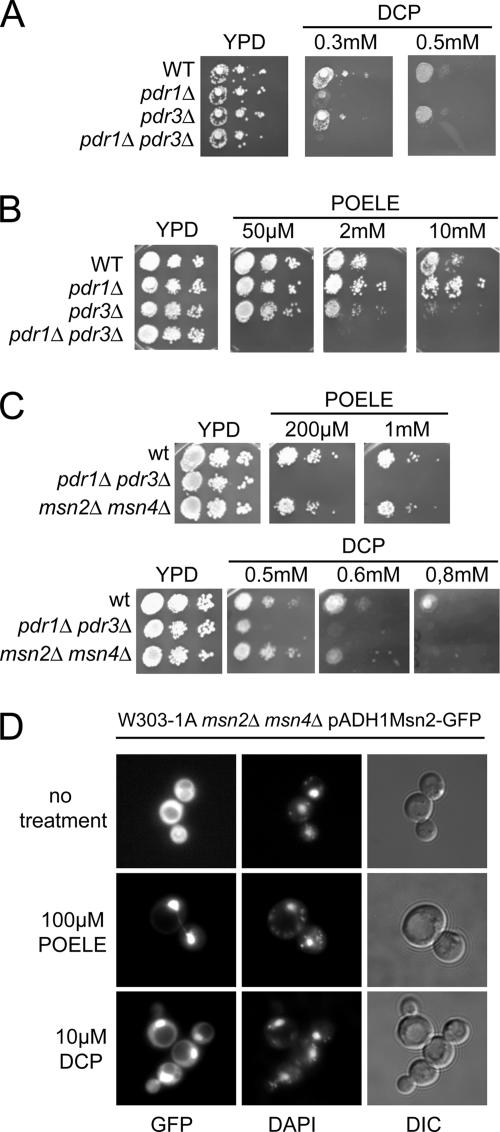
The activation of PDR and general stress response by DCP and nonionic detergents raised the question about a role for the corresponding transcription factors Pdr1/Pdr3 and Msn2/Msn4 in physiological responses to membrane lipid perturbance. We thus analyzed growth properties of pdr1Δ pdr3Δ and msn2Δ msn4Δ mutant cells on plates to investigate whether the respective transcription factors are required for resistance against high concentrations of DCP or POELE. Serial dilutions of isogenic wild-type, naΔ1 (pdr1Δ), naΔ3 (pdr3Δ), and naΔ1Δ3 (pdr1Δ pdr3Δ) cells were spotted onto YPD plates containing increasing amounts of these compounds. Growth of wild-type and pdr3Δ cells was unaffected by up to 0.5 mM DCP. However, growth of pdr1Δ cells was inhibited by 0.3 mM DCP, and pdr1Δ pdr3Δ double mutants cells failed to grow at this concentration (Figure 2A). A plasmid carrying the PDR1 gene restored DCP resistance in both naΔ1 and naΔ1Δ3 strains (data not shown). Exposure of pdr1Δ and pdr3Δ mutants to POELE produced a different growth pattern (Figure 2B). pdr1Δpdr3Δ cells were highly sensitive and unable to grow in the presence of 50 μM POELE. Surprisingly, wild-type cells were unaffected by concentrations of up to 10 mM POELE. At this concentration, growth of pdr3Δ cells was almost absent compared with the wild type, whereas pdr1Δ mutants grew even slightly better than wild-type cells (Figure 2B). This indicated that Pdr1 and Pdr3 are functionally nonredundant under these conditions. Similarly, the pdr1Δ pdr3Δ double mutant was also hypersensitive to other nonionic detergents such as NP-40 and Triton X-100 (data not shown). By contrast, the msn2Δmsn4Δ double mutant failed to show any growth defects on POELE-containing plates. Like wild-type cells, the msn2Δmsn4Δ cells were able to grow in the presence of 1 mM POELE. However, at high concentrations of DCP (0.6 and 0.8 mM), msn2Δmsn4Δ cells displayed increased sensitivities when compared with the wild-type control (Figure 2C).
Figure 2.
POELE and DCP activate Pdr1/Pdr3 and Msn2/Msn4, leading to DCP resistance. (A) FY1679-28c (WT), naΔ1 (pdr1Δ), naΔ3 (pdr3Δ), and naΔ1Δ3 (pdr1Δ pdr3Δ) cells were grown on YPD to OD600 1.0. Then, they were diluted to an OD600 of 0.2, 0.02, and 0.002 and spotted on plates containing increasing amounts of DCP. Cell growth was inspected after 3 d. (B) FY1679-28c (WT), naΔ1 (pdr1Δ), naΔ3 (pdr3Δ), and naΔ1Δ3 (pdr1Δ pdr3Δ) cells were grown in YPD to an OD600 of 1.0 and diluted to an OD600 of 0.4, 0.04, and 0.004, and then they were spotted onto plates containing increasing concentrations of POELE. Cell growth was inspected after 3 d. (C) W303-1A (WT), YYA100 (pdr1Δ pdr3Δ), and msn2Δ msn4Δ growing in the exponential growth phase were diluted to an OD600 of 0.4, 0.04, and 0.004, and then they were spotted onto YPD plates containing the indicated concentrations of either DCP or POELE. Plates were inspected after 3-d incubation at 30°C. (D) W303-1A msn2Δ msn4Δ cells were transformed with pADH1-Msn2-GFP to express Msn2-GFP. Msn2-GFP fluorescence was monitored in living exponentially growing cells treated with 0.1 mM POELE or 0.3 mM DCP. 4,6-Diamidino-2-phenylindole staining was used to visualize nuclear DNA.
To confirm that DCP and POELE are directly activating Msn2, we used a Msn2-GFP reporter construct driven by the ADH1 promoter (pAMG) (Görner et al., 1998). The intracellular localization of Msn2 strongly reflects the stress status of cells. Strain W303-1A msn2Δ msn4Δ expressing Msn2-GFP was grown to the exponential phase in SC medium and treated with 0.1 mM POELE or 0.3 mM DCP. Msn2-GFP fluorescence was monitored in living cells without fixation (Figure 2D). Although Msn2-GFP was distributed throughout the cytoplasm in untreated cells, POELE caused a rapid and almost complete nuclear translocation of Msn2-GFP within 1 min. Likewise, DCP treatment also resulted in enhanced nuclear accumulation of Msn2-GFP, although this response was less pronounced compared with POELE stress. To investigate the activation of Msn2 target genes, we also measured Ctt1 catalase activities to monitor STRE-dependent transcription, because catalase T is almost exclusively regulated by Msn2/Msn4. Catalase activity increased ∼10-fold in cells treated with 0.1 mM POELE for 45 min, demonstrating a rapid activation of Msn2/Msn4 (data not shown). Membrane perturbance may also trigger a high osmolarity response. We therefore tested a hog1Δ strain under the same experimental conditions, but we failed to detect a difference to the wild-type control (data not shown). Together, these results indicate that the general stress mediator Msn2 is activated by both DCP and POELE. Furthermore, Msn2 and Msn4 contribute to DCP resistance, whereas Pdr1 and Pdr3 are required for resistance to both DCP and the nonionic detergent POELE.
Lack of Pdr5 and Pdr15 Causes Hypersensitivity to DCP and Nonionic Detergents
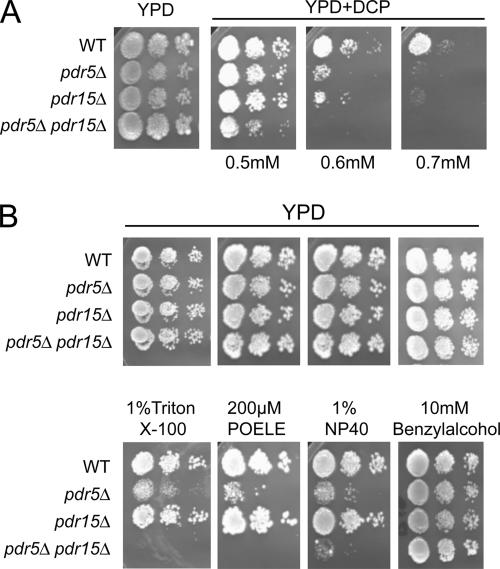
The multidrug transporter Pdr5 is a major target of Pdr1 and Pdr3, and it was demonstrated to confer tolerance to elevated 2,4-D levels (Teixeira and Sa-Correia, 2002; Teixeira et al., 2006). Our results suggest the importance of Pdr1/Pdr3 and their target ABC drug efflux pumps for resistance against other membrane-active agents as well. In normal cells, Pdr5 levels are highly dependent on the presence of Pdr1/Pdr3. Interestingly, we have shown that Pdr5 and Pdr15 show a complimentary expression regulation in different growth phases and carbon sources (Mamnun et al., 2004); Pdr15 is the closest functional homologue to Pdr5 and is also regulated by Pdr1/Pdr3 and by the general stress regulators Msn2/Msn4. To test the effect of these closely related transporters on DCP tolerance, we spotted wild-type cells (W303-1A) and cells lacking PDR5 (NRY201), PDR15 (NRY212), or both transporters (NRY227) onto YPD plates containing different amounts of DCP (Figure 3A). At 0.5 mM DCP, the double deletion strain showed severely reduced growth, whereas the single deletion strains lacking PDR5 or PDR15 displayed no growth phenotypes compared with wild-type cells. At higher concentrations of DCP (0.6 and 0.7 mM), pdr5Δ pdr15Δ double mutants failed to grow, and growth of pdr5Δ and pdr15Δ single deletion strains was impaired. Similar results were also obtained using another set of isogenic pdr5Δ, pdr15Δ, and pdr5Δ pdr15Δ deletion strains in a different genetic background (YPH499) (data not shown). Notably, absence of other closely related ABC transporters such as Pdr10, Snq2, or Yor1 did not cause any hypersensitivity (data not shown), suggesting a central role for Pdr5 and Pdr15 in resistance to DCP.
Figure 3.
Deletion of PDR5 and PDR15 causes increased sensitivity to detergents. W303-1A (WT), NRY201 (pdr5Δ), NRY212 (pdr15Δ), and NRY227 (pdr5Δ pdr15Δ) were grown to exponential phase, diluted to an OD600 of 0.4, 0.04, and 0.004 and spotted onto YPD plates containing different concentrations of DCP (A) and 1% Triton X-100, 200 μM POELE, 1% NP-40, or 10 mM benzyl alcohol (B). Growth was inspected after 3 d, and for DCP plates, after 4 d.
The same isogenic set of strains was also spotted onto YPD plates containing different concentrations of POELE, NP-40, Triton X-100, or benzyl alcohol, which was reported to increase bulk membrane fluidity in rat liver cells (Gordon et al., 1980). Cells lacking Pdr5 or both Pdr5 and Pdr15, displayed increased sensitivities to all three nonionic detergents (Figure 3B). Exposure to 1% Triton X-100, 200 μM POELE, or 1% NP-40 strongly impaired growth of pdr5Δ cells compared with wild-type or pdr15Δ cells. Nevertheless, the double mutant pdr5Δ pdr15Δ showed stronger sensitivity to the detergents (Figure 3B), revealing a contribution of Pdr15 to the resistance phenotype. The double mutant pdr5Δ pdr15Δ was unable to grow on 0.1% Triton X-100 or 50 μM POELE (data not shown), and it showed strong growth defects on 1% NP-40. None of the deletion strains displayed any hypersensitivity to benzyl alcohol. Hence, both ABC transporters must contribute to detergent resistance in an additive effect. Failure to detect growth defects of a pdr15Δ strain suggests that Pdr5 shares a redundant function and thus functionally masks Pdr15 activity due to its high level of expression compared with Pdr5 (Wolfger et al., 2004).
PDR5 and PDR15 Are Strongly Induced by Various Membrane-damaging Agents
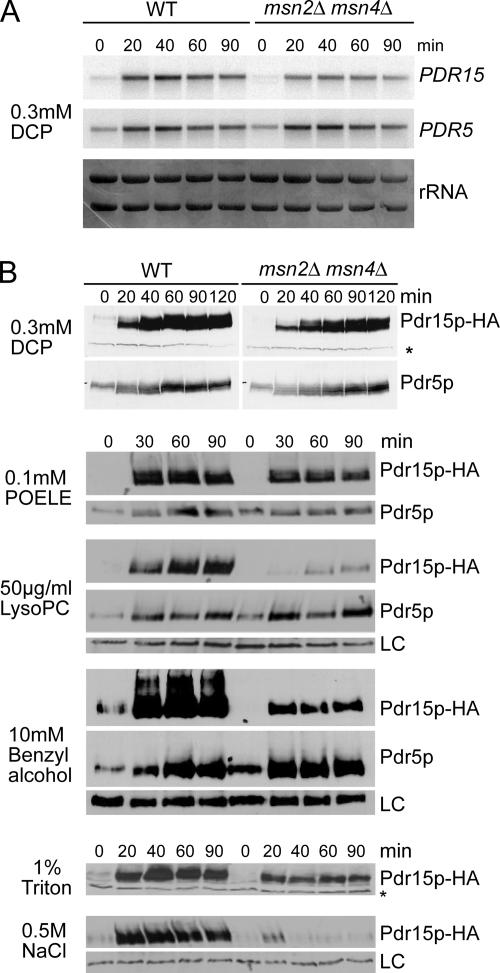
To reconfirm that transcription of PDR5 and PDR15 is induced by DCP, exponentially growing W303-1A cells were exposed to 0.3 mM DCP for up to 90 min and mRNA levels of PDR5 and PDR15 were detected by Northern blotting. PDR15 mRNA levels were very low in exponentially growing cells, but they strongly increased within the first 20 min of DCP treatment, and they remained high over the next 90-min period (Figure 4A). Because we have recently shown that PDR15 expression is regulated by Msn2/Msn4, which mediate strong Pdr15 induction in response to several adverse conditions (Wolfger et al., 2004), a strain lacking both Msn2 and Msn4 was included in this experiment. Surprisingly, absence of Msn2/Msn4 had only minor influence on the expression of PDR15 under these conditions, indicating a strong requirement for Pdr1/Pdr3. Similarly, PDR5 mRNA also rapidly increased in the presence of DCP independent of Msn2/Msn4.
Figure 4.
Membrane-damaging agents and detergents strongly induce Pdr5 and Pdr15. (A) W303-1A and msn2Δ msn4Δ cells were grown to an OD600 of 1.0 in YPD and treated with 0.3 mM DCP. Samples were taken before stress and after the indicated time. PDR5 and PDR15 mRNAs were detected by Northern blotting. rRNA bands confirm equal sample loading. (B) YHW4 (WT) and YHW5 (msn2Δ msn4Δ) cells were grown to OD600 of 1.0 in YPD and treated with 0.3 mM DCP, 100 μM POELE, 10 mM benzyl alcohol, 50 μg/ml lysoPC, 1% Triton X-100, or 0.5 M NaCl. Protein extracts were prepared and Pdr5 Pdr15-3HA levels detected by immunoblotting using anti-Pdr5 and anti-HA 16B12 antibodies, respectively. A cross-reaction of the anti-HA antibody (LC) was used to verify equal protein loading. Samples were taken before stress treatment and at the indicated times.
To asses whether DCP stress also modulates corresponding protein levels of both transporters, we used the strains YHW4 and YHW5 (msn2Δ msn4Δ), expressing genomic levels of epitope-tagged Pdr15-3HA (Figure 4B). Protein levels of Pdr15-3HA steeply increased within 30 min of DCP treatment and remained high over at least 2 h. Equally high amounts of Pdr15-3HA in msn2Δ msn4Δ cells confirmed that induction by DCP bypasses Msn2 and Msn4. Notably, Pdr5 levels, although already very high in unstressed, exponentially growing cells (Mamnun et al., 2004), still showed a robust increase in response to DCP treatment. The results show that both ABC transporters Pdr5 and Pdr15 are required for DCP resistance and highly induced by this substance independently of Msn2/Msn4.
Because Pdr5 and Pdr15 are also required for resistance to various detergents, we tested whether both transporters are also induced by these substances or by other membrane-damaging agents. Strains YHW4 (WT) and YHW5 (msn2Δ msn4Δ) expressing Pdr15-3HA were treated with 0.1 mM POELE, 1% Triton X-100, 10 mM benzyl alcohol, or 50 μg/ml lysoPC (Figure 4B). Triton X-100 and POELE caused a rapid and massive increase of Pdr15-HA levels in the wild-type strain. As shown for DCP, this induction was largely independent of Msn2/Msn4, although msn2Δ msn4Δ cells lacking both transcription factors expressed slightly reduced levels of Pdr15. A similar effect was also observed for benzyl alcohol (Figure 4B), where cells lacking Msn2/Msn4 still displayed a strong induction of Pdr15, although levels were reduced in general. A significant contribution of Msn2/Msn4 was only observed for the induction of Pdr15 in response to lysoPC, a natural membrane lipid breakdown product with strong detergent activity. The high basal expression of Pdr5 was only slightly increased by all three compounds. As a positive control, we used the strong induction of Pdr15 by 0.5 M NaCl (Wolfger et al., 2004), which strictly required the presence of Msn2/Msn4 (Figure 4B). Therefore, compounds with the potential to interfere with or impair membrane lipid integrity cause a high and mainly Msn2/Msn4-independent but Pdr1/Pdr3-depenedent induction of Pdr5 and Pdr15.
Pdr1 and Pdr3 Respond to Membrane Stress by Induction of PDR5 and PDR15
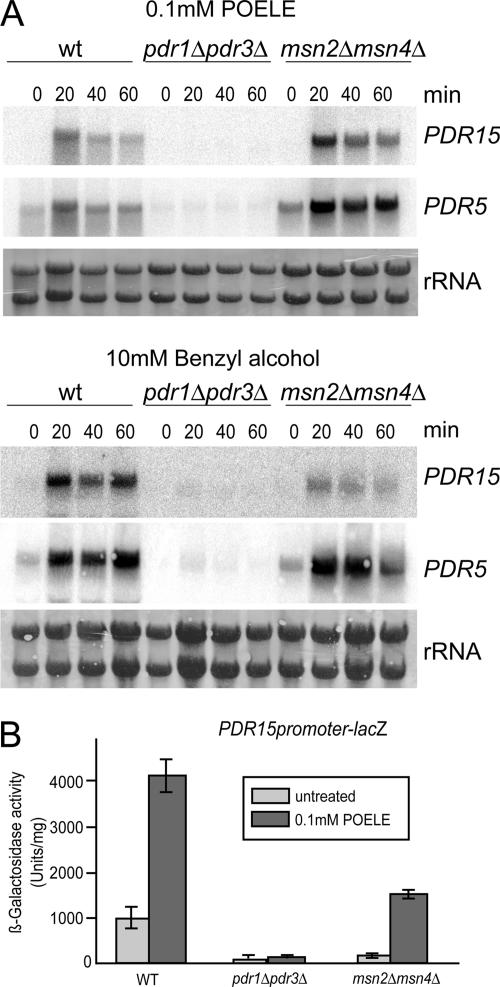
Both PDR5 and PDR15 harbor several PDREs in their promoters, which place them under transcriptional control of Pdr1 and Pdr3. Because the microarray analyses showed a strong induction of PDR genes, we tested whether Pdr1 and Pdr3 are required for induction of PDR15 expression during membrane stress. We used an isogenic set of strains lacking Pdr1 and Pdr3 (YYA100, pdr1Δ pdr3Δ) and cells devoid of both Msn2 and Msn4 (msn2Δ msn4Δ). Treatment of exponentially growing cells with 0.1 mM POELE or 10 mM benzyl alcohol (Figure 5A) resulted in a strong and rapid increase of PDR15 and PDR5 mRNA levels in both wild-type and msn2Δ msn4Δ cells. Strikingly, this induction was absent in cells lacking Pdr1 and Pdr3.
Figure 5.
Induction of PDR15 and PDR5 is mainly dependent on Pdr1 and Pdr3. (A) W303-1A (WT), YA100 (pdr1Δ pdr3Δ), and msn2Δ msn4Δ growing in exponential growth phase were treated with either 100 μM POELE or 10 mM benzyl alcohol. Samples were taken before stress treatment and at the indicated times. Expression of PDR5 and PDR15 was analyzed by Northern blotting by using the appropriate probes. rRNA bands serve as loading control. (B) W303-1A (WT), YYA100 (pdr1Δ pdr3Δ), and msn2Δ msn4Δ carrying the plasmid pHW-15z were grown overnight on selective medium, shifted to YPD, and grown to the exponential phase for 4 h. Samples were either left untreated (light gray) or treated with 0.1 mM POELE (dark gray) for 90 min. β-Galactosidase activities were determined in crude cell free extracts. All experiments were carried out in triplicates.
The levels of PDR5 mRNA were also strongly elevated in response to both stress agents, and this induction seemed entirely dependent on Pdr1/Pdr3. To verify that PDR15 induction is dependent on the transcriptional activation of its promoter, we used a PDR15 promoter-lacZ reporter construct. β-Galactosidase assays were performed in cell-free extracts of W303-1A, YYA100 (pdr1Δ pdr3Δ) and W303 msn2Δ msn4Δ cells treated with 0.1 mM POELE. Reporter activity increased ∼fourfold in wild-type and msn2Δ msn4Δ cells in response to POELE, although overall β-galactosidase activities were significantly lower in the mutant, confirming the results of the previous experiments. Importantly, pdr1Δ pdr3Δ cells displayed a very low reporter activity, which was not increased in response to POELE treatment. This confirms that Pdr1 and Pdr3, but not Msn2 and Msn4, are the main regulators for the POELE-induced transcriptional activation of PDR15 (Figure 5B).
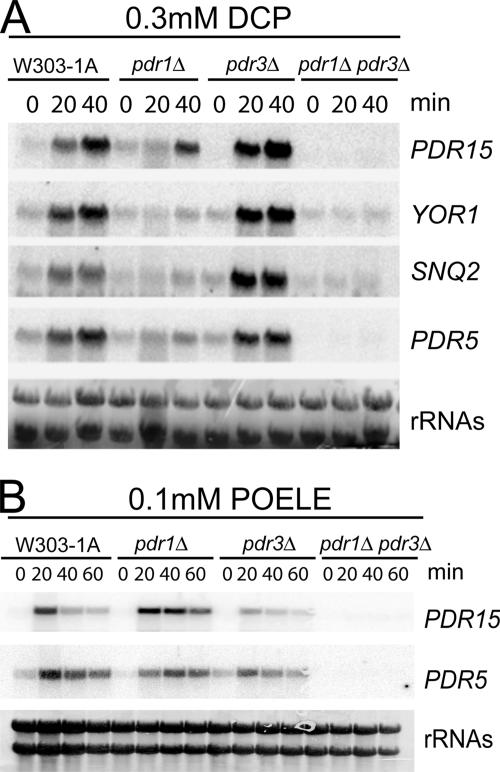
The viability assays in the presence of DCP suggested distinct roles for Pdr1 and Pdr3. Thus, we investigated the influence of Pdr1/Pdr3 on the transcriptional response of several PDRE-regulated target genes by using Northern blotting (Figure 6). The isogenic strains FY1679-28c (WT), naΔ1 (pdr1Δ), naΔ3 (pdr3Δ), and naΔ1Δ3 (pdr1Δ pdr3Δ) were treated with either 0.3 mM DCP or 0.1 mM POELE, and RNA from these cells was used for Northern analysis (Figure 6). Although a lack of PDR3 did not cause any effects on PDR15, PDR5, SNQ2, and YOR1 mRNA levels after DCP treatment, induction was nearly absent in pdr1Δ cells and in pdr1Δ pdr3Δ double mutants. This identified Pdr1 as the main mediator of DCP stress response, which is in full agreement with the results from growth assays on agar plates (Figure 2A).
Figure 6.
Differential induction of Pdr1 and Pdr3 target genes by DCP and POELE. (A) FY1679-28c (WT), naΔ1 (pdr1Δ), naΔ3 (pdr3Δ), and naΔ1Δ3 (pdr1Δ pdr3Δ) cells were grown on YPD to an OD600 of 1.0 and treated with 0.3 mM DCP for 20 and 40 min. Samples were taken before and during stress treatment and PDR15, YOR1, PDR5, and SNQ2 expression was analyzed by Northern blotting. rRNA bands confirm equal sample loading. (B) FY1679-28c (WT), naΔ1 (pdr1Δ), naΔ3 (pdr3Δ), and naΔ1Δ3 (pdr1Δ pdr3Δ) cells were grown in YPD to an OD600 of 1.0 and treated with 100 μM POELE for 20, 40, and 60 min. Samples were taken before and during stress treatment and PDR15 and PDR5 mRNA levels were analyzed by Northern blotting. rRNA bands confirm equal sample loading.
In contrast to DCP, Pdr1 and Pdr3 seem to have overlapping functions in the response to POELE. Wild-type, pdr1Δ, and pdr3Δ cells showed a robust increase of PDR15 and PDR5 mRNA upon treatment with 0.1 mM POELE (Figure 6B) or 1% Triton X-100 (data not shown), whereas the response was only abolished in the pdr1Δ pdr3Δ double mutant. This indicated that Pdr1 and Pdr3, despite sharing overlapping functions, can also mediate distinct stress responses depending on the nature of the inducing cues.
Detergent Treatment Activates the PDREs in the Target Gene Promoter
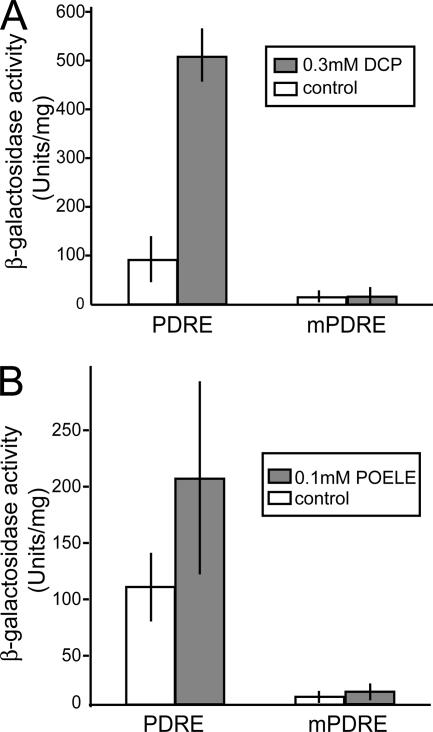
Because Pdr1 and Pdr3 regulate transcription via PDREs in their target promoters, we next investigated whether PDREs are sufficient to mediate transcriptional activation. Hence, we used an otherwise unregulated heterologous promoter containing ectopic PDREs, and we tested the activation by DCP or POELE. Cells carrying plasmids with PDREs (pDK52) or mutated PDREs (pDK53) inserted into a disabled CYC1 promoter driving the lacZ reporter gene (Katzmann et al., 1996) were grown in YPD to exponential growth phase, and then they were treated with either 0.3 mM DCP or 0.1 mM POELE for 2 h (Figure 7). Both substances activated the heterologous promoter carrying the functional but not the mutated PDREs, as shown by the activity of the β-galactosidase reporter. DCP resulted in a fivefold induction, whereas POELE increased β-galactosidase activity ∼twofold (Figure 7). Similar results were obtained from independent transformants and in parallel experiments, showing that both stress conditions directly act through PDRE elements.
Figure 7.
DCP and POELE activate transcription through the PDRE. W303-1A cells carrying pDK52 (PDRE) or pDK53 (mPDRE) were shifted from selective medium to YPD and grown in exponential phase for 4 h. Samples were taken before and after 2-h treatment with 0.3 mM DCP (A) or 100 μM POELE (B), and β-galactosidase activity was determined in protein extracts. Experiments were carried out at least in triplicates.
Pdr5 and Pdr15 May Regulate Plasma Membrane Asymmetry
Pdr5 and the more distantly related ABC transporter Yor1 have been proposed to function as outward-directed lipid translocases (“floppases”) for plasma membrane PE (Kean et al., 1997; Decottignies et al., 1998; Pomorski et al., 2003). Pdr15 has not been examined in this context so far. We, therefore, examined the phospholipid composition and asymmetric plasma membrane phosphatidylethanolamine distribution of wild-type, pdr5Δ, pdr15Δ, and pdr5Δ pdr15Δ cells. However, no significant differences in overall phospholipid composition were observed (data not shown), because PE distribution was unaffected in pdr5Δ and pdr15Δ cells, with wild-type cells exposing 1.4 ± 0.2% of total phosphatidylethanolamine and pdr5Δ and pdr15Δ cells exposing 1.4 ± 0.3 and 1.4 ± 0.1%, respectively. The pdr5Δ pdr15Δ double mutant strain exhibited 1.7-fold increased exposure (2.4 ± 0.1%), suggesting that Pdr5 and Pdr15 may influence or modulate the transbilayer lipid distribution of certain plasma membrane phospholipids.
DISCUSSION
The transcription factors Pdr1 and Pdr3 drive expression of a set of genes, including several ABC transporters (Wolfger et al., 2001; Moye-Rowley, 2003), conferring tolerance to many unrelated cytotoxic substances. Long-term drug treatment promotes selection of constitutive hyperactive alleles of these factors, giving rise to PDR phenotypes (Anderson et al., 2003), mainly due to hyperinduction of several detoxifying membrane efflux pumps of the ABC transporter family (Carvajal et al., 1997; Wolfger et al., 2001). Thus, mutations in Pdr1 and Pdr3 may represent an emergency route to PDR under selective pressure (Anderson et al., 2003). Apart from their function as constitutive transcription activators, Pdr1 and Pdr3 have also been reported to become activated by certain compounds such as the phenolic herbicide 2,4-D (Teixeira and Sa-Correia, 2002) or fluphenazine (Fardeau et al., 2007). Genome-wide expression profiling showed that Pdr1/Pdr3 trigger a rapid and transient transcriptional reprogramming quite similar to other environmental stress responses such as heat shock (Teixeira et al., 2006; Fardeau et al., 2007).
The substances driving Pdr1/Pdr3-dependent regulation are structurally and functionally unrelated (Supplemental Figure S1), raising the question about the trigger of Pdr1/Pdr3 activation. In the absence of upstream regulatory pathways, the mechanism of Pdr1/Pdr3 activation might be similar to those of related binuclear Zn(II)2Cys6 zinc cluster transcription factors, several of which may be directly regulated by binding activating compounds or pathway intermediates (Sellick and Reece, 2005). The binding of a ligand could cause structural changes that trigger activation, as suggested for Put3 (Des Etages et al., 2001) and other zinc cluster transcription factors (Phelps et al., 2006). Likewise, we have recently also demonstrated that a rapid conformational change of the weak acid stress regulator War1 is triggered by its stress-induced activation (Schüller et al., unpublished data). One common feature shared between the PDR-inducing compounds is their hydrophobicity. The solubility of these compounds in water is generally low, with 2 mg/l for benomyl (log P of 23.4–32.0), 31 mg/l for fluphenazine (log P of 4.552), and 0.89g/l for 2,4-D (log P of 1.934). We also investigated the transcriptional response to two apparently chemically unrelated compounds, DCP (a phenolic substance with a solubility in water 4g/l; log P of 3.06) and the nonionic detergent POELE (log P of 14). Both compounds caused a rapid induction of a wide range of genes regulated by several stress response pathways. From 147 and 193 genes induced by DCP (>1.6) and POELE (>2.0), respectively, most if not all overlap with those genes induced by 2,4-D (Teixeira et al., 2006) and fluphenazine (Fardeau et al., 2007). Genes depending on Pdr1/Pdr3 were the most prominent group of highly induced genes. Deletion of both factors completely abolished this induction (Figure 1A) by DCP. Furthermore, a PDRE-driven reporter system demonstrates that PDREs are sufficient to mediate the DCP and PEOLE response. This confirms that both substances indeed increase the transcriptional activity of the PDR regulators most likely by activating Pdr1 and/or Pdr3.
In addition to PDR-related genes, DCP and POELE treatment generate signals for the activation of other transcriptional regulators. T-Profiler analysis revealed induction of genes containing motifs for Hsf1, Rpn4, Crz1, Ino4, and Msn2/Msn4 binding (Lee et al., 2002; Boorsma et al., 2005). Similar results have been obtained for fluphenazine, which is activating Msn2- and Crz1-regulated genes, whereas benomyl caused induction of oxidative stress response via Yap1 (Fardeau et al., 2007). Msn2/Msn4 have been previously implicated in signaling membrane damage (Moskvina et al., 1999) and 2,4-D (Simoes et al., 2003). Increase of PDR gene transcription is more pronounced upon DCP treatment, whereas the POELE response is broader. Induction by DCP of Msn2/Msn4-regulated genes is reduced by the absence of Pdr1/Pdr3, pointing to a cross talk between PDR and general stress response.
The relevance of a transcriptional response is determined at the level of target genes. We therefore investigated the physiological relevance of the PDR response and its properties concerning the regulation on single PDR target genes. Deletion of Msn2/Msn4 results in some sensitivity to DCP. However, lack of Pdr1 and Pdr3 causes a pronounced hypersensitivity of cells to DCP and nonionic detergents. From their targets, in particular the ABC transporter Pdr5 and to a lesser extent Pdr15, both are required for growth in the presence of nonionic detergents such as Triton X-100, NP-40, lysoPC, or POELE. This shows that Pdr1/Pdr3 and distinct target genes are involved in counteracting the adverse effect of membrane lipid detergents, and it also points to a possible function of Pdr15 and Pdr5 in counteracting or sensing membrane damage.
Pdr1 and Pdr3 might recognize and directly bind membrane-active compounds or soluble breakdown products. A similar situation might exist in the suspected sterol transporters Aus1 and Pdr11 controlled by the transcription factors Ucp2 and Ecm22, which are probably binding sterol biosynthesis intermediates, thereby modulating sterol homeostasis (Wilcox et al., 2002). Control of Pdr1 and Pdr3 activity might also be similar to that of the related transcription factors Pip2 and Oaf1, which are required for peroxisome biogenesis and possibly bind fatty acids or related metabolic intermediates (Rottensteiner et al., 1996; Baumgartner et al., 1999).
If membrane damage or perturbance is activating Pdr1 and Pdr3, the question of the inducing signal remains open. Because both factors are constitutively localized in the nucleus (Mamnun et al., 2002), a signal generated from lipid disturbances at the plasma membrane has to be transduced to the nucleus. The activating signal could also come from an indirect source such as the mitochondria. 2,4-D induces apoptosis in human lymphocytes via mitochondrial damage (Kaioumova et al., 2001b). Strikingly, Pdr3 has indeed been implicated in sensing and signaling mitochondrial damage to some of its target genes (Devaux et al., 2002). In fact, Pdr3, but not Pdr1, is specifically and rapidly activated by mitochondrial malfunction (Hallström and Moye-Rowley, 2000), suggesting a distinct mechanism for the Pdr1-dependent activation by DCP. Pdr1 and/or Pdr3 could also respond to lipid degradation products generated during the repair process from membrane lipid constituents of the plasma membrane. This idea is supported by the induction of Pdr1/Pdr3-regulated genes after treatment with the toxic lipid peroxidation product linoleic acid hydroperoxide (Alic et al., 2003).
Interestingly, Pdr5 and other as yet unidentified Pdr1/Pdr3 target genes have been proposed to act as phospholipid translocators (floppases and “flippases”) in the plasma membrane (Mahé et al., 1996a; Kean et al., 1997; Kihara and Igarashi, 2004). Recent reports along this line show that Rsb1, a putative translocator of sphingoid long-chain bases is regulated by Pdr1/Pdr3 (Panwar and Moye-Rowley, 2006). Notably, other Pdr1/Pdr3 targets also seem to regulate sphingolipid synthesis (Hallström et al., 2001) and sterol trafficking (Vik and Rine, 2001). Our results from this study also show that PE exposure is increased in pdr5 pdr15 double mutants. This suggests that Pdr5 and Pdr15 could be involved in maintaining the membrane lipid bilayer distribution in the plasma membrane. Our observations (Rockwell, unpublished data) implicate the Pdr10 ABC transporter as a further regulator of plasma membrane asymmetry.
Together, our work provides compelling evidence for a function of Pdr1 and Pdr3, and their target genes Pdr5 and Pdr15, in regulating membrane lipid organization in addition to their roles for detoxification. The ABC transporters might mediate removal of breakdown products of membrane phospholipids or short-lived oxidized lipids, which might otherwise cause membrane malfunctions. This function may also explain the high turnover and endocytosis rate of Pdr5 (Egner et al., 1995), because removal of membrane-damaging lipids into the extracellular space through a classical transport step would not remove the “drug,” whereas endocytosis and subsequent vacuolar delivery could. Thus, certain yeast ABC transporters such as Pdr5 and Pdr15 may guard and maintain membrane bilayer function by binding and removing unwanted or toxic lipids, many of which have features similar to hydrophobic xenobiotics. As our results demonstrate, this system is controlled by the pleiotropic drug resistance regulators Pdr1 and Pdr3, which seem to become activated by hydrophobic drug-like compounds and membrane-active detergents, connecting phenomena such as drug resistance with membrane lipid stress response.
Supplementary Material
ACKNOWLEDGMENTS
We thank our colleagues Scott Moye-Rowley and David Katzman for the gift of PDRE reporter plasmids. This work was supported by a grant from the “Fonds zur Förderung der wissenschaftlichen Forschung” (P-15934-B08), and by the EC FP6 Marie Curie Training Network “Flippases” (MCRTN-CT-2004-005330).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-06-0610) on September 19, 2007.
REFERENCES
- Alic N., Higgins V. J., Pichova A., Breitenbach M., Dawes I. W. Lipid hydroperoxides activate the mitogen-activated protein kinase Mpk1p in Saccharomyces cerevisiae. J. Biol. Chem. 2003;278:41849–41855. doi: 10.1074/jbc.M307760200. [DOI] [PubMed] [Google Scholar]
- Anderson J. B., Sirjusingh C., Parsons A. B., Boone C., Wickens C., Cowen L. E., Kohn L. M. Mode of selection and experimental evolution of antifungal drug resistance in Saccharomyces cerevisiae. Genetics. 2003;163:1287–1298. doi: 10.1093/genetics/163.4.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgartner U., Hamilton B., Piskacek M., Ruis H., Rottensteiner H. Functional analysis of the Zn(2)Cys(6) transcription factors Oaf1p and Pip2p. Different roles in fatty acid induction of beta-oxidation in Saccharomyces cerevisiae. J. Biol. Chem. 1999;274:22208–22216. doi: 10.1074/jbc.274.32.22208. [DOI] [PubMed] [Google Scholar]
- Boorsma A., Foat B. C., Vis D., Klis F., Bussemaker H. J. T-profiler: scoring the activity of predefined groups of genes using gene expression data. Nucleic Acids Res. 2005;33:W592–W595. doi: 10.1093/nar/gki484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazma A., et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat. Genet. 2001;29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- Carvajal E., van den Hazel H. B., Cybularz-Kolaczkowska A., Balzi E., Goffeau A. Molecular and phenotypic characterization of yeast PDR1 mutants that show hyperactive transcription of various ABC multidrug transporter genes. Mol. Gen. Genet. 1997;256:406–415. doi: 10.1007/s004380050584. [DOI] [PubMed] [Google Scholar]
- Chang S. C., Heacock P. N., Clancey C. J., Dowhan W. The PEL1 gene (renamed PGS1) encodes the phosphatidylglycero-phosphate synthase of Saccharomyces cerevisiae. J. Biol. Chem. 1998;273:9829–9836. doi: 10.1074/jbc.273.16.9829. [DOI] [PubMed] [Google Scholar]
- Chua G., Morris Q. D., Sopko R., Robinson M. D., Ryan O., Chan E. T., Frey B. J., Andrews B. J., Boone C., Hughes T. R. Identifying transcription factor functions and targets by phenotypic activation. Proc. Natl. Acad. Sci. USA. 2006;103:12045–12050. doi: 10.1073/pnas.0605140103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decottignies A., Grant A. M., Nichols J. W., de Wet H., McIntosh D. B., Goffeau A. ATPase and multidrug transport activities of the overexpressed yeast ABC protein Yor1p. J. Biol. Chem. 1998;273:12612–12622. doi: 10.1074/jbc.273.20.12612. [DOI] [PubMed] [Google Scholar]
- Decottignies A., Kolaczkowski M., Balzi E., Goffeau A. Solubilization and characterization of the overexpressed PDR5 multidrug resistance nucleotide triphosphatase of yeast. J. Biol. Chem. 1994;269:12797–12803. [PubMed] [Google Scholar]
- Delaveau T., Delahodde A., Carvajal E., Subik J., Jacq C. PDR3, a new yeast regulatory gene, is homologous to PDR1 and controls the multidrug resistance phenomenon. Mol. Gen. Genet. 1994;244:501–511. doi: 10.1007/BF00583901. [DOI] [PubMed] [Google Scholar]
- DeRisi J., van den Hazel B., Marc P., Balzi E., Brown P., Jacq C., Goffeau A. Genome microarray analysis of transcriptional activation in multidrug resistance yeast mutants. FEBS Lett. 2000;470:156–160. doi: 10.1016/s0014-5793(00)01294-1. [DOI] [PubMed] [Google Scholar]
- Des Etages S. A., Saxena D., Huang H. L., Falvey D. A., Barber D., Brandriss M. C. Conformational changes play a role in regulating the activity of the proline utilization pathway-specific regulator in Saccharomyces cerevisiae. Mol. Microbiol. 2001;40:890–899. doi: 10.1046/j.1365-2958.2001.02432.x. [DOI] [PubMed] [Google Scholar]
- Devaux F., Carvajal E., Moye-Rowley S., Jacq C. Genome-wide studies on the nuclear PDR3-controlled response to mitochondrial dysfunction in yeast. FEBS Lett. 2002;515:25–28. doi: 10.1016/s0014-5793(02)02387-6. [DOI] [PubMed] [Google Scholar]
- Devaux F., Marc P., Bouchoux C., Delaveau T., Hikkel I., Potier M. C., Jacq C. An artificial transcription activator mimics the genome-wide properties of the yeast Pdr1 transcription factor. EMBO Rep. 2001;2:493–498. doi: 10.1093/embo-reports/kve114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egner R., Mahé Y., Pandjaitan R., Kuchler K. Endocytosis and vacuolar degradation of the plasma membrane-localized Pdr5 ATP-binding cassette multidrug transporter in Saccharomyces cerevisiae. Mol. Cell. Biol. 1995;15:5879–5887. doi: 10.1128/mcb.15.11.5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis L.B., Roe D., Wackett L.P. The University of Minnesota Biocatalysis/Biodegradation Database: the first decade. Nucleic Acids Res. 2006;34:D517–D521. doi: 10.1093/nar/gkj076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esko J. D., Raetz C. R. Autoradiographic detection of animal cell membrane mutants altered in phosphatidylcholine synthesis. Proc. Natl. Acad. Sci. USA. 1980;77:5192–5196. doi: 10.1073/pnas.77.9.5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fardeau V., Lelandais G., Oldfield A., Salin H., Lemoine S., Garcia M., Tanty V., Le Crom S., Jacq C., Devaux F. The central role of PDR1 in the foundation of yeast drug resistance. J. Biol. Chem. 2007;282:5063–5074. doi: 10.1074/jbc.M610197200. [DOI] [PubMed] [Google Scholar]
- Flynn P. J., Reece R. J. Activation of transcription by metabolic intermediates of the pyrimidine biosynthetic pathway. Mol. Cell. Biol. 1999;19:882–888. doi: 10.1128/mcb.19.1.882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch A. P., Spellman P. T., Kao C. M., Carmel-Harel O., Eisen M. B., Storz G., Botstein D., Brown P. O. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordesky S. E., Marinetti G. V., Segel G. B. The interaction of 1-fluoro-2,4-dinitrobenzene with amino-phospholipids in membranes of intact erythrocytes, modified erythrocytes, and erythrocytes ghosts. J. Membr. Biol. 1973;14:229–242. doi: 10.1007/BF01868080. [DOI] [PubMed] [Google Scholar]
- Gordon L. M., Sauerheber R. D., Esgate J. A., Dipple I., Marchmont R. J., Houslay M. D. The increase in bilayer fluidity of rat liver plasma membranes achieved by the local anesthetic benzyl alcohol affects the activity of intrinsic membrane enzymes. J. Biol. Chem. 1980;255:4519–4527. [PubMed] [Google Scholar]
- Görner W., Durchschlag E., Martinez-Pastor M. T., Estruch F., Ammerer G., Hamilton B., Ruis H., Schüller C. Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes Dev. 1998;12:586–597. doi: 10.1101/gad.12.4.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., Kohlhaw G. B. Regulation of transcription in mammalian cells by yeast Leu3p and externally supplied inducer. FEBS Lett. 1996;390:191–195. doi: 10.1016/0014-5793(96)00653-9. [DOI] [PubMed] [Google Scholar]
- Hallström T. C., Lambert L., Schorling S., Balzi E., Goffeau A., Moye-Rowley W. S. Coordinate control of sphingolipid biosynthesis and multidrug resistance in Saccharomyces cerevisiae. J. Biol. Chem. 2001;276:23674–23680. doi: 10.1074/jbc.M101568200. [DOI] [PubMed] [Google Scholar]
- Hallström T. C., Moye-Rowley W. S. Divergent transcriptional control of multidrug resistance genes in Saccharomyces cerevisiae. J. Biol. Chem. 1998;273:2098–2104. doi: 10.1074/jbc.273.4.2098. [DOI] [PubMed] [Google Scholar]
- Hallström T. C., Moye-Rowley W. S. Multiple signals from dysfunctional mitochondria activate the pleiotropic drug resistance pathway in Saccharomyces cerevisiae. J. Biol. Chem. 2000;275:37347–37356. doi: 10.1074/jbc.M007338200. [DOI] [PubMed] [Google Scholar]
- Kaioumova D., Kaioumov F., Opelz G., Susal C. Toxic effects of the herbicide 2,4-dichlorophenoxyacetic acid on lymphoid organs of the rat. Chemosphere. 2001a;43:801–805. doi: 10.1016/s0045-6535(00)00436-7. [DOI] [PubMed] [Google Scholar]
- Kaioumova D., Susal C., Opelz G. Induction of apoptosis in human lymphocytes by the herbicide 2,4-dichlorophenoxyacetic acid. Hum. Immunol. 2001b;62:64–74. doi: 10.1016/s0198-8859(00)00229-9. [DOI] [PubMed] [Google Scholar]
- Kaiser C., Michaelis S., Mitchell A. A Laboratory Course Manual, Cold Spring Harbor. New York: Cold Spring Harbor Laboratory Press; 1994. Methods in Yeast Genetics. [Google Scholar]
- Katzmann D. J., Burnett P. E., Golin J., Mahé Y., Moye-Rowley W. S. Transcriptional control of the yeast PDR5 gene by the PDR3 gene product. Mol. Cell. Biol. 1994;14:4653–4661. doi: 10.1128/mcb.14.7.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzmann D. J., Hallström T. C., Mahé Y., Moye-Rowley W. S. Multiple Pdr1p/Pdr3p binding sites are essential for normal expression of the ATP binding cassette transporter protein-encoding gene PDR5. J. Biol. Chem. 1996;271:23049–23054. doi: 10.1074/jbc.271.38.23049. [DOI] [PubMed] [Google Scholar]
- Kean L. S., Grant A. M., Angeletti C., Mahe Y., Kuchler K., Fuller R. S., Nichols J. W. Plasma membrane translocation of fluorescent-labeled phosphatidylethanolamine is controlled by transcription regulators, PDR1 and PDR3. J. Cell Biol. 1997;138:255–270. doi: 10.1083/jcb.138.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara A., Igarashi Y. Cross talk between sphingolipids and glycerophospholipids in the establishment of plasma membrane asymmetry. Mol. Biol. Cell. 2004;15:4949–4959. doi: 10.1091/mbc.E04-06-0458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T. I., et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- MacPherson S., Larochelle M., Turcotte B. A fungal family of transcriptional regulators: the zinc cluster proteins. Microbiol. Mol. Biol. Rev. 2006;70:583–604. doi: 10.1128/MMBR.00015-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahé Y., Lemoine Y., Kuchler K. The ATP binding cassette transporters Pdr5 and Snq2 of Saccharomyces cerevisiae can mediate transport of steroids in vivo. J. Biol. Chem. 1996a;271:25167–25172. doi: 10.1074/jbc.271.41.25167. [DOI] [PubMed] [Google Scholar]
- Mahé Y., Parle-McDermott A., Nourani A., Delahodde A., Lamprecht A., Kuchler K. The ATP-binding cassette multidrug transporter Snq2 of Saccharomyces cerevisiae: a novel target for the transcription factors Pdr1 and Pdr3. Mol. Microbiol. 1996b;20:109–117. doi: 10.1111/j.1365-2958.1996.tb02493.x. [DOI] [PubMed] [Google Scholar]
- Mamnun Y. M., Pandjaitan R., Mahe Y., Delahodde A., Kuchler K. The yeast zinc finger regulators Pdr1p and Pdr3p control pleiotropic drug resistance (PDR) as homo- and heterodimers in vivo. Mol. Microbiol. 2002;46:1429–1440. doi: 10.1046/j.1365-2958.2002.03262.x. [DOI] [PubMed] [Google Scholar]
- Mamnun Y. M., Schüller C., Kuchler K. Expression regulation of the yeast PDR5 ATP-binding cassette (ABC) transporter suggests a role in cellular detoxification during the exponential growth phase. FEBS Lett. 2004;559:111–117. doi: 10.1016/S0014-5793(04)00046-8. [DOI] [PubMed] [Google Scholar]
- Moskvina E., Imre E. M., Ruis H. Stress factors acting at the level of the plasma membrane induce transcription via the stress response element (STRE) of the yeast Saccharomyces cerevisiae. Mol. Microbiol. 1999;32:1263–1272. doi: 10.1046/j.1365-2958.1999.01438.x. [DOI] [PubMed] [Google Scholar]
- Moye-Rowley W. S. Transcriptional control of multidrug resistance in the yeast Saccharomyces cerevisisae. Prog. Nucleic Acids Res. Mol. Biol. 2003;73:251–279. doi: 10.1016/s0079-6603(03)01008-0. [DOI] [PubMed] [Google Scholar]
- Panwar S. L., Moye-Rowley W. S. Long chain base tolerance in Saccharomyces cerevisiae is induced by retrograde signals from the mitochondria. J. Biol. Chem. 2006;281:6376–6384. doi: 10.1074/jbc.M512115200. [DOI] [PubMed] [Google Scholar]
- Phelps C., Gburcik V., Suslova E., Dudek P., Forafonov F., Bot N., MacLean M., Fagan R. J., Picard D. Fungi and animals may share a common ancestor to nuclear receptors. Proc. Natl. Acad. Sci. USA. 2006;103:7077–7081. doi: 10.1073/pnas.0510080103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomorski T., Lombardi R., Riezman H., Devaux P. F., van Meer G., Holthuis J. C. Drs2p-related P-type ATPases Dnf1p and Dnf2p are required for phospholipid translocation across the yeast plasma membrane and serve a role in endocytosis. Mol. Biol. Cell. 2003;14:1240–1254. doi: 10.1091/mbc.E02-08-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reece R. J., Beynon L., Holden S., Hughes A. D., Rebora K., Sellick C. A. Nutrient-regulated gene expression in eukaryotes. Biochem. Soc. Symp. 2006;73:85–96. doi: 10.1042/bss0730085. [DOI] [PubMed] [Google Scholar]
- Rottensteiner H., Kal A. J., Filipits M., Binder M., Hamilton B., Tabak H. F., Ruis H. Pip2p: a transcriptional regulator of peroxisome proliferation in the yeast Saccharomyces cerevisiae. EMBO J. 1996;15:2924–2934. [PMC free article] [PubMed] [Google Scholar]
- Sellick C. A., Reece R. J. Eukaryotic transcription factors as direct nutrient sensors. Trends Biochem. Sci. 2005;30:405–412. doi: 10.1016/j.tibs.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Siegmund A., Grant A., Angeletti C., Malone L., Nichols J. W., Rudolph H. K. Loss of Drs2p does not abolish transfer of fluorescence-labeled phospholipids across the plasma membrane of Saccharomyces cerevisiae. J. Biol. Chem. 1998;273:34399–34405. doi: 10.1074/jbc.273.51.34399. [DOI] [PubMed] [Google Scholar]
- Simoes T., Teixeira M., Fernandes C.A.R., Sa′-Correia I. Adaptation of Saccharomyces cerevisiae to the herbicide 2,4-dichlorophenoxyacetic acid, mediated by Msn2p- and Msn4p-regulated genes important role of SPI1. Appl. Environ. Microbiol. 2003;69:4019–4028. doi: 10.1128/AEM.69.7.4019-4028.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipos G., Kuchler K. Fungal ATP-binding cassette (ABC) transporters in drug resistance & detoxification. Curr. Drug Targets. 2006;7:471–481. doi: 10.2174/138945006776359403. [DOI] [PubMed] [Google Scholar]
- Suwalsky M., Benites M., Villena F., Aguilar F., Sotomayor C. P. Interaction of 2,4-dichlorophenoxyacetic acid (2,4-D) with cell and model membranes. Biochim. Biophys. Acta. 1996;1285:267–276. doi: 10.1016/s0005-2736(96)00173-3. [DOI] [PubMed] [Google Scholar]
- Teixeira M. C., Fernandes A. R., Mira N. P., Becker J. D., Sa-Correia I. Early transcriptional response of Saccharomyces cerevisiae to stress imposed by the herbicide 2,4-dichlorophenoxyacetic acid. FEMS Yeast Res. 2006;6:230–248. doi: 10.1111/j.1567-1364.2006.00041.x. [DOI] [PubMed] [Google Scholar]
- Teixeira M.C., Sa-Correia I. Saccharomyces cerevisiae resistance to chlorinated phenoxyacetic acid herbicides involves Pdr1p-mediated transcriptional activation of TPO1 and PDR5 genes. Biochem. Biophys. Res. Commun. 2002;292:530–537. doi: 10.1006/bbrc.2002.6691. [DOI] [PubMed] [Google Scholar]
- Teixeira M. C., Telo J. P., Duarte N. F., Sa-Correia I. The herbicide 2,4-dichlorophenoxyacetic acid induces the generation of free-radicals and associated oxidative stress responses in yeast. Biochem. Biophys. Res. Commun. 2004;324:1101–1107. doi: 10.1016/j.bbrc.2004.09.158. [DOI] [PubMed] [Google Scholar]
- Tuschl H., Schwab C. Cytotoxic effects of the herbicide 2,4-dichlorophenoxyacetic acid in HepG2 cells. Food Chem. Toxicol. 2003;41:385–393. doi: 10.1016/s0278-6915(02)00238-7. [DOI] [PubMed] [Google Scholar]
- Vik A., Rine J. Upc2p and Ecm22p, dual regulators of sterol biosynthesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 2001;21:6395–6405. doi: 10.1128/MCB.21.19.6395-6405.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Zheng F., Holmberg S., Kohlhaw G. B. Yeast transcriptional regulator Leu3p. Self-masking, specificity of masking, and evidence for regulation by the intracellular level of Leu3p. J. Biol. Chem. 1999;274:19017–19024. doi: 10.1074/jbc.274.27.19017. [DOI] [PubMed] [Google Scholar]
- Wilcox L. J., Balderes D. A., Wharton B., Tinkelenberg A. H., Rao G., Sturley S. L. Transcriptional profiling identifies two members of the ATP-binding cassette transporter superfamily required for sterol uptake in yeast. J. Biol. Chem. 2002;277:32466–32472. doi: 10.1074/jbc.M204707200. [DOI] [PubMed] [Google Scholar]
- Wolfger H., Mahé Y., Parle-McDermott A., Delahodde A., Kuchler K. The yeast ATP binding cassette (ABC) protein genes PDR10 and PDR15 are novel targets for the Pdr1 and Pdr3 transcriptional regulators. FEBS Lett. 1997;418:269–274. doi: 10.1016/s0014-5793(97)01382-3. [DOI] [PubMed] [Google Scholar]
- Wolfger H., Mamnun Y. M., Kuchler K. Fungal ABC proteins: pleiotropic drug resistance, stress response and cellular detoxification. Res. Microbiol. 2001;152:375–389. doi: 10.1016/s0923-2508(01)01209-8. [DOI] [PubMed] [Google Scholar]
- Wolfger H., Mamnun Y. M., Kuchler K. The yeast Pdr15p ATP-binding cassette (ABC) protein is a general stress response factor implicated in cellular detoxification. J. Biol. Chem. 2004;279:11593–11599. doi: 10.1074/jbc.M311282200. [DOI] [PubMed] [Google Scholar]
- Yano K., Fukasawa T. Galactose-dependent reversible interaction of Gal3p with Gal80p in the induction pathway of Gal4p-activated genes of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 1997;94:1721–1726. doi: 10.1073/pnas.94.5.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto H., Saltsman K., Gasch A. P., Li H. X., Ogawa N., Botstein D., Brown P. O., Cyert M. S. Genome-wide analysis of gene expression regulated by the calcineurin/Crz1p signaling pathway in Saccharomyces cerevisiae. J. Biol. Chem. 2002;277:31079–31088. doi: 10.1074/jbc.M202718200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.