Abstract
Recent reports have indicated the participation of tight junction (TJ) proteins in the regulation of gene expression and cell proliferation. Here, we have studied the role of zona occludens (ZO)-2, a TJ peripheral protein, in the regulation of cyclin D1 transcription. We found that ZO-2 down-regulates cyclin D1 transcription in a dose-dependent manner. To understand how ZO-2 represses cyclin D1 promoter activity, we used deletion analyses and found that ZO-2 negatively regulates cyclin D1 transcription via an E box and that it diminishes cell proliferation. Because ZO-2 does not associate directly with DNA, electrophoretic mobility shift assay and chromatin immunoprecipitation (ChIP) assay were used to identify the transcription factors mediating the ZO-2–repressive effect. c-Myc was found to bind the E box present in the cyclin D1 promoter, and the overexpression of c-Myc augmented the inhibition generated by ZO-2 transfection. The presence of ZO-2 and c-Myc in the same complex was further demonstrated by immunoprecipitation. ChIP and reporter gene assays using histone deacetylases (HDACs) inhibitors demonstrated that HDACs are necessary for ZO-2 repression and that HDAC1 is recruited to the E box. We conclude that ZO-2 down-regulates cyclin D1 transcription by interacting with the c-Myc/E box element and by recruiting HDAC1.
INTRODUCTION
Zona occludens (ZO)-2 is a peripheral tight junction (TJ) protein that belongs to the membrane associated guanylate kinases (MAGUK) protein family. ZO-2, and the other epithelial cell ZO proteins named ZO-1 and ZO-3, possess the basic MAGUK core of Src homology (SH)3 and GuK domains, and they harbor three postsynaptic density 95/disc-large/zona occludens (PDZ) repeats. These proteins additionally present a carboxy-terminal region, which in the case of ZO-1 and ZO-2 contains a proline-rich module (Gonzalez-Mariscal et al., 2000). The subcellular localization of ZO-2 is strongly sensitive to the state of cell–cell contacts displayed by the monolayer. Thus, in confluent monolayers ZO-2 is concentrated at the TJ, whereas in sparse cultures it is enriched at the nucleus and cell–cell contacts. Shuttling between the nucleus and the plasma membrane is possible due to the presence of several functional nuclear localization and exportation signals in the sequence of the protein (Jaramillo et al., 2004; Gonzalez-Mariscal et al., 2006).
ZO-2 associates with the nuclear matrix (Jaramillo et al., 2004), and it displays a speckled nuclear distribution that partially colocalizes with that of the pre-mRNA splicing protein SC-35 (Islas et al., 2002). ZO-2 interacts with scaffold attachment factor B, a chromatin component involved in the assembly of transcriptosome complexes (Traweger et al., 2002), and with the transcription factors Jun, Fos, and CCAAT/enhancer-binding protein (C/EBP). ZO-2 has further been shown to inhibit the transcription of a reporter gene with a promoter under the control of activator protein (AP)-1 sites (Betanzos et al., 2004).
The canonical role of TJs is to act as a gate that regulates the passage of ions and molecules through the paracellular pathway and to function as a fence that maintains the polarized distribution of lipids and proteins in the plasma membrane. However, in recent years, the role of TJs as regulators of cell proliferation and differentiation has started to emerge. For example, ZO-1, another MAGUK protein of the TJ, has been found to associate through its SH3 domain to ZONAB, a Y-box transcription factor that stimulates cell proliferation (Balda and Matter, 2000; Balda et al., 2003; Sourisseau et al., 2006).
Several studies suggest that ZO-2 functions as a tumor suppressor in epithelial cells. First, ZO proteins are homologous to the product of the lethal disc large-1 tumor suppressor gene of Drosophila (Willott et al., 1993). Second, ZO-2 expression is down-regulated in breast and pancreas human carcinomas (Chlenski et al., 1999, 2000), and third, the overexpression of ZO-2 suppresses transformation by the oncogenic determinants E4 from adenovirus type 9, Ras V12, and the polyomavirus middle T protein (Glaunsinger et al., 2001).
In this study, we have further pursued the participation of ZO-2 in gene transcription, and due to the emerging role of TJ proteins in cell proliferation, we have analyzed the impact of ZO-2 on the transcription of cyclin D1. Cyclin D assembles with CDK4/6 in early G1. Accumulation of this complex leads to the activation of the kinases that phosphorylate and inactivate the tumor suppressor retinoblastoma, a necessary step for cell cycle progression through G1-to-S phases (Harbour and Dean, 2000). The expression levels of cyclin D1 have been shown to be rate limiting in cellular proliferation induced by a variety of stimuli (Ohtsubo and Roberts, 1993; Quelle et al., 1993; Albanese et al., 1995; Watanabe et al., 1996), and an increased expression of cyclin D1 has been observed in several tumors (Wang et al., 1994; Arber et al., 1996; Zhang et al., 1997; Albanese et al., 1999).
Here, we have analyzed cyclin D1 promoter activity under the influence of ZO-2. We observed that ZO-2 represses cyclin D1 transcription in a manner that is dependent on a region localized between nucleotides −694 to −550 in the promoter, which harbors a typical E box. c-Myc and HDAC1 bind to this E box and in a complex with ZO-2 inhibit cyclin D1 transcription, resulting in a diminished cell proliferation rate. Together, these data support the idea of ZO-2 as a participant in the transcriptional control of cyclin D1, a key cell cycle regulator.
MATERIALS AND METHODS
Cell Culture
Epithelial Madin Darby canine kidney (MDCK) cultures (MDCK, CCL34) were obtained from the American Type Culture Collection (Manassas, VA). Cells between the 60th and 90th passage were grown at 37°C, 5% CO2 atmosphere with DMEM (Sigma-Aldrich, St. Louis, MO) containing 10% fetal bovine serum, 50 μg/ml gentamicin, and 100 IU/ml penicillin. Cells were harvested with trypsin-EDTA (Invitrogen, Carlsbad, CA) and plated on multiwell plates for further assays, seeding 2.5 × 104 cells/cm2.
Plasmids
Full-length ZO-2 introduced into the cytomegalovirus (CMV) expression plasmid pGW1 (pGW1-HA-ZO-2) was kindly provided by Dr. Ronald Javier (Baylor College of Medicine, Houston, TX). pCB6-ZO-1myc was kindly provided by Dr. Maria Susana Balda (Institute of Ophthalmology, University College of London, London, United Kingdom). pHMyc3E plasmid contains the human c-Myc cDNA, exons 1-3 (Jones and Cole, 1987), modified in an LTR/pBR322 plasmid (denoted only as pBR322), and it was a kind gift of Dr. Patricio Gariglio (Cinvestav, Mexico). The luciferase reporter gene under the control of the human cyclin D1 promoter (Herber et al., 1994) (pXP2-CD1) and its mutated versions at 12-O-tetradecanoylphorbol 13-acetate (TPA)-response element (TRE/AP-1) (pXP2-CD1mTRE) and CRE sites (pXP2-CD1mCRE), and the pRSV-β-Gal plasmid with a Rous sarcoma virus promoter and β-galactosidase reporter (Thierry et al., 1992), were kind gifts of Dr. Francoise Thierry (Institute Pasteur, Paris, France). pXP2-CD1 was also used to generate three deletion constructs that eliminate the E2F site by using the following pairs of restriction enzymes: BstxI and BlpI (p-694ΔCycD1), MscI and PmlI (p-550ΔCycD1), and BstxI and DsaI (p-492ΔCycD1) (see map in Figure 4B). For pSoxCAT construction, the double-stranded Sox10 oligonucleotide from Myelin Basic Protein Gene Promoter (Wei et al., 2004) was cloned upstream of the simian virus 40 promoter (pCAT-promoter vector; Promega, Madison, WI). All plasmids were purified using the QIAGEN plasmid purification kit (QIAGEN, Valencia, CA), verified by restriction and complete automated sequence analyses.
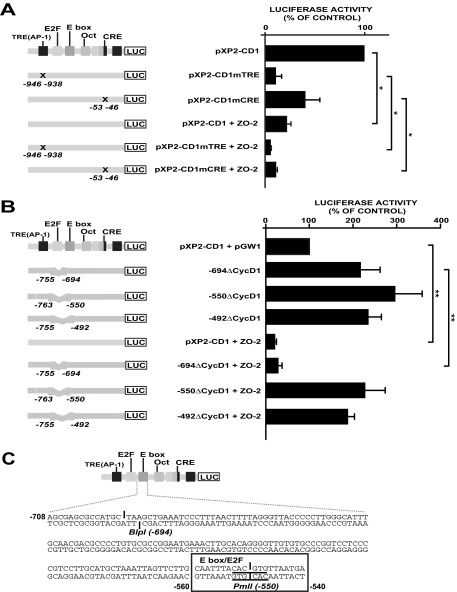
Figure 4.
ZO-2 repression is mediated by a putative E2F/E box located in the cyclin D1 promoter. (A) The effects of punctual mutations on cyclin D1 promoter, depicted as X in the TRE(AP-1) (change TGAGTCA→CGGTACC) and CRE (change TAACGTCA→TAAAGCTT) sites, were monitored by reporter assays done under a control condition and under the influence of ZO-2 overexpression. MDCK cells were transfected with 1 μg of pXP2-CD1, the TRE(AP-1) and CRE mutant constructs or one of these constructs together with 2 μg of pGW1-HA-ZO-2. Relative luciferase activities are expressed as a percentage of the activity of the control (pXP2-CD1 alone). (B) Cyclin D1 promoter constructs with diverse deletions over the E2F binding site were analyzed under a control condition and under the influence of ZO-2 overexpression. MDCK cells were transfected with 1 μg of pXP2-CD1 or the E2F binding site deletions constructs alone or together with 2 μg of pGW1-HA-ZO-2. Relative luciferase activities are expressed as a percentage of the activity of pXP2-CD1/pGW1 taken as the control. *p < 0.05, **p < 0.001, Student's t test. At the left side, mutagenesis sites and deletions are indicated. (C) In silico analysis of the cyclin D1 promoter sequence, located between −708 and −540, confirms the presence of an E box and reveals a putative E2F binding site. Chosen restriction enzymes are indicated.
Transient Transfections and Reporter Assays
Transient transfections were performed in sparse MDCK cultures by using Lipofectamine Plus (Invitrogen) or Lipofectamine 2000 (Invitrogen) protocol, under the manufacturer's conditions. Amounts used of each reporter plasmid and expression vector are indicated in each figure. Cells were incubated with the mixture for 3 h. After that, cells were incubated for a further 48 h with fresh medium to allow recovery. They were subsequently harvested and processed for reporter measurement. Luciferase assays were performed with the Luciferase assay system (Promega). Briefly, protein lysates were obtained as follows. Cells were harvested in cold phosphate-buffered saline (PBS) and lysed with two freeze-thawing cycles and resuspended in 100 μl of reporter lysis buffer. Equal amounts of protein lysates (∼70 μg) were incubated with luciferase assay reagent. Light detection was performed in a FluoroSkan Ascent FL 374 (Thermo Electron, Waltham, MA). For chloramphenicol acetyltransferase (CAT) assays, normalized protein lysates (in lysis buffer, up to ≈80 μg) were incubated with 0.25 μCi of [14C]chloramphenicol (50 mCi/mmol; GE Healthcare, Chalfont St. Giles, United Kingdom) and 0.8 mmol/l acetyl-CoA (Sigma-Aldrich) at 37°C. Acetylated forms were separated by thin layer chromatography and quantified using a Typhoon optical scanner (GE Healthcare). CAT activities were expressed as the acetylated fraction corrected for the activity recorded from empty-vector–transfected control. For β-galactosidase assays, normalized amounts of protein lysates were used to determine β-galactosidase activity by using 200 μl of O-nitrophenyl β-d-galactopyranoside solution (4 mg/ml; Sigma-Aldrich) as substrate in 500 μl of Z-buffer (50 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, and 1 mM MgCl2, pH 7.0). After incubation for 16 h at 37°C, the reaction was stopped by adding 200 μl of 1 M Na2CO3, and the absorbance of the reaction product was read at 410 nm using an enzyme-linked immunosorbent assay (ELISA) plate reader (Opsys MR; Dynex Technologies, Chantilly, VA).
For chromatin immunoprecipitation (ChIP) assays, MDCK cells were electroporated with pGW1-derived plasmids. For this purpose, cells were harvested and washed in PBS by low-speed centrifugation. Cells (1 × 107) were suspended in 400 μl of basal DMEM medium containing 20 μg of pGW1-HA-ZO-2 plasmid. Electroporation was carried out at room temperature using single voltage pulse (250 mV; 960 μF) with a Gene-Pulser electroporator (Bio-Rad, Hercules, CA). Cells were then plated into 15-cm cell culture dishes. Two days later, cells were collected for the ChIP assay.
Cell Proliferation Assays
DNA synthesis was assessed using [3H]thymidine; cells were seeded in six-well plates, transfected with indicated vectors, and incubated for 12–96 h with 1 μCi/ml [3H]thymidine. The incorporation was stopped with cold PBS, precipitation with 5% trichloroacetic acid, and cell lysis with 0.4 M NaOH. After 1 h of gentle agitation at room temperature, 15 μl of each sample were taken for protein quantification, and the rest was solubilized in 25 μl of 10% acetic acid and 2 ml of scintillation liquid. Samples were counted in a beta-scintillation counter (LS6000SC, Beckman Coulter, Fullerton, CA).
For 3-(4, 5-dimethylthiazolyl-2)-2,5-diphenyltetrazolium bromide (Sigma-Aldrich) MTT reduction assay (Mosmann, 1983), cells were seeded on 12-well plates in 500 μl of culture media and incubated at 37°C in 5% CO2. They were analyzed in quadruplicates at 48 h. Then, 40 μl of MTT labeling mixture was added to each well, and the samples were incubated 4 h at 37°C in 5% CO2. An isopropanol:HCl solution was added to lyse the cells and to solubilize the colored crystals. The optical density of the samples was determined at 630 nm using an ELISA plate reader Opsys MR (Dynex Technologies).
Wound Healing Assay
For wound healing assays, MDCK cells (4 × 104/cm2) were seeded in eight-well glass slide chambers (Nalge Nunc International, Rochester, NY). After 3 h, cells were transfected with the pGW1 or pGW1-HA-ZO-2 plasmids (5 μg/individual chamber), or they were transfected with small interfering RNAs (siRNAs) (negative control [NC] or siRNA ZO-2; 10 nM). Then, cells were incubated for 48 h to reach confluence. Wounds were introduced to the MDCK-transfected confluent monolayers with a yellow pipette tip to create a cleared line. Monolayers were washed twice with PBS to remove detached cells, and they were incubated with complete media. The wound closure process was photographed, focusing always in the same region every 3 h after wounding (12 h total) and by using an Olympus IX250 microscope (Olympus, Melville, NY). The number of pixels in the denuded area for at least four independent chambers was calculated using the Adobe Photoshop software (Adobe Systems, San Jose, CA).
ChIP Assays
Sparse MDCK cells were transiently transfected with 20 μg of pGW1-HA-ZO-2. Forty-eight hours posttransfection, cells were treated with 1% formaldehyde for 20 min to cross-link protein–DNA and protein–protein complexes. Chromatin was broken by sonication. The chromatin fragments were immunoprecipitated with 4 μg of the following antibodies: ZO-2 (catalog no. 71-1400; Zymed Laboratories, South San Francisco, CA), hemagglutinin (HA) (catalog no. 7500; Zymed Laboratories), H3Ac (catalog no. 07-353; Upstate Biotechnology), H3 (catalog no. 06-755; Upstate Biotechnology, Lake Placid, NY), HDAC1 (catalog no. sc-7872; Santa Cruz Biotechnology, Santa Cruz, CA) and c-Myc (catalog no. sc-788l Santa Cruz Biotechnology). Preimmune serum and no antibody were used as negative controls. Immunoprecipitates were washed with several buffers and treated with RNase for 1 h, incubated overnight at 65°C to remove cross-linking, and then treated with proteinase K for 2 h. DNA was purified by phenol/chloroform extraction and ethanol precipitation. DNA samples were analyzed by radioactive polymerase chain reactions (PCRs) by using the following pairs of primers: 1) a pair that amplified the E box region present in the cyclin D1 promoter (ChIPEboxD1S 5′-CGAGCGCATGCTAGGCTGAA-3′ and ChIPEboxD1AS 5′-CTCCGCGTCCACATCTTTCA-3′); 2) a second set of primers that amplified the TRE site present in the cyclin D1 promoter (ChIPAP-1D1S 5′-AACAAAACCAATTAAAAAGCTT-3′ and ChIPAP-1D1AS 5′-ATTTCCTTCATCCTGTCCTC-3′); and 3) a third pair of oligonucleotides that amplified part of the first exon of cyclin D1 gene and which were used as a negative control (dogD1S 5′-CTGGCCATGAACTACCTGGA-3′ and CNS 5′-CCAGCAGTTCGTCGGGCCG-3′). PCR reactions were performed at annealing temperature of 59°C for 30 cycles. The products were analyzed in 5% polyacrylamide gels and visualized in a Typhoon optical scanner (GE Healthcare) or exposed to an autoradiographic film.
Transfection of siRNA
To obtain an effective and highly specific knockdown of ZO-2, we used a Stealth RNA interference (Invitrogen) that was previously shown by us to effectively silence ZO-2 expression (Hernandez et al., 2007). The target sequence for dog ZO-2 siRNA was 5′-CCGACUAUGAUUAUCAUUCCUCAAA-3′.
ZO-2 siRNA (10 nM) was transfected into MDCK cells by using Lipofectamine 2000. For reporter assays, the siRNA transfection was performed 24 h before the transfection of the pXP2-CD1 promoter, and the cells were harvested 48 h after the second transfection. For reverse transcription (RT)-PCR, immunodetection and wound healing assays, the transfection was realized as described above, and cultures were harvested 72 h after transfection (Hernandez et al., 2007). RNA extraction and RT-PCR were performed as described below.
Electrophoretic Mobility Shift Assays
Nuclear extracts were prepared as described previously (Lopez-Bayghen et al., 1996). All buffers contained a protease inhibitors cocktail to prevent nuclear factor proteolysis. Protein concentration was measured by the Bradford method (Bradford, 1976). Nuclear extracts (∼10 μg) from sparse MDCK cultures were incubated on ice with 500 ng of poly[dI-dC] as nonspecific competitor (GE Healthcare) and 2 ng of the following 32P-end–labeled double-stranded oligonucleotides: E box/E2F, 5′-CTAGCAATTTACACGTGTTAATGA-3′ and c-Myc, 5′-CTAGGAAGCAGACCACGTGGTCTGCTTCC-3′.
The reaction mixtures were incubated for 20 min on ice, electrophoresed in 7.8% polyacrylamide gels by using a low ionic strength 0.5× Tris borate-EDTA buffer. The gels were then dried and exposed to an autoradiographic film. For competitive analysis, the reactions mixtures were incubated before the addition of labeled DNA, with 100 ng of the following unlabeled competitor oligonucleotides: mMyc, 5′-GGAAGCAGACCACGGAGTCTGCTTCC-3′; E2F, 5′-CTAGATTTAAGTTTCGCGCCCTTTCTCAA-3′; mE2F, 5′-ATTTAAGTTTCGATCCCTTTCTCAA-3′; and AP-1, 5′-CTAGTTCCGGCTGAGTCATCAAGC-3′.
RT-PCR
Total RNA was isolated from MDCK cells transiently transfected with different amounts of pGW1-HA-ZO-2 or the empty vector (pGW1). Total RNA was extracted from cells using the TRIzol method (Invitrogen). One microgram of total RNA was used in each analysis. RT-PCR reactions were performed using the Enhanced Avian HS RT-PCR kit (Sigma-Aldrich). The reverse-transcription step was performed at 42°C for 50 min. Amplification conditions for both cyclin D1 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were as follows: after an initial 94°C incubation cycle for 3 min, reactions were amplified for 35 cycles at 94°C for 1 min, 55°C for 2 min, and 72°C for 3 min. The reactions were then incubated at 72°C for 10 min. PCR amplification products were separated on a 1% agarose gel and visualized by ethidium bromide staining. Primers used to amplify the cyclin D1 gene were the following: dogD1S, 5′-CTGGCCATGAACTACCTGGA-3′ and dogD1AS, 5′-GGAAGTGCTCGATGAAGTCG-3′. As control, we used the following specific primers to the GAPDH gene (Hsu et al., 1993): primer 1, 5′-CATCTCTGCCCCCTCTGCTGA-3′ and primer 2, 5′-GGATGACCTTGCCCACAGCCT-3′.
Data were quantified by scanning the bands and the optical densities of cyclin D1 were normalized to the GAPDH signal. Data shown are representative of at least three independent experiments.
Protein Blotting
Total extracts from MDCK cells that overexpress ZO-2 or c-Myc, and nuclear extracts from sparse MDCK monolayers (initial seeding 2.5 × 104 cells/cm2) were separated by SDS-polyacrylamide gel electrophoresis and transferred into polyvinylidene difluoride membranes (GE Healthcare). Proteins were identified with polyclonal antibodies against ZO-2 (Zymed Laboratories; dilution 1:1000), cyclin D1 (catalog no. sc-717; Santa Cruz Biotechnology; dilution 1:1000), c-Myc (Santa Cruz Biotechnology; dilution 1:1000), HDAC1 (Santa Cruz Biotechnology; dilution 1:1000), and anti-actin (a kind gift of Dr. José Manuel Hernández, Cinvestav, Mexico; dilution 1:50). Peroxidase-conjugated goat immunoglobulin (Ig)G against rabbit IgG or against mouse IgG (catalog no. 62-6120 and 62-6520, respectively, Zymed Laboratories; dilution 1:3000) were used as secondary antibodies, followed by a chemiluminescence detection system ECL+ Plus (GE Healthcare).
Treatments for HDAC Inhibition
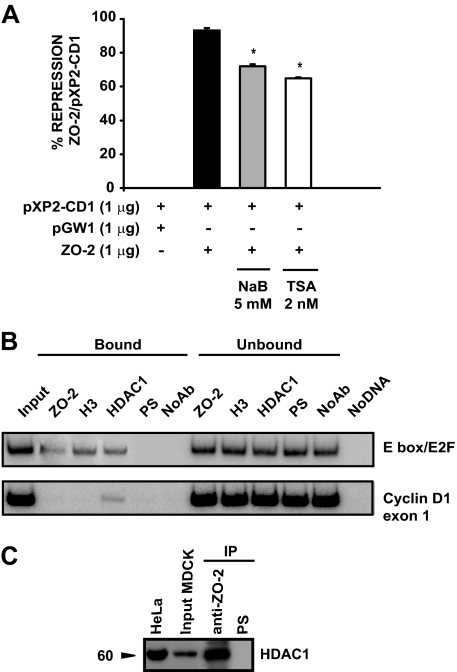
For evaluating HDAC activity, cells transfected with pXP2-CD1 and pGW1-ZO-2 were treated with the HDAC inhibitors trichostatin A (TSA; 2 nM) and sodium butyrate (NaB; 5 mM) for 12 h before harvesting.
Immunoprecipitation
Nuclear extracts from MDCK cells that overexpressed ZO-2 were homogenized in radioimmunoprecipitation assay (RIPA) buffer. The cell debris of both preparations was removed by centrifugation at 13,000 rpm for 5 min. Then, 500 μg of protein was used for each immunoprecipitation, with 1 μg of the antibody against ZO-2 (catalog no. 37-4700, Zymed Laboratories). The cell lysates were preabsorbed with protein A coupled to agarose for 25 min at 4°C. Agarose-coupled antibodies were incubated overnight at 4°C with protein samples. Agarose beads were washed three times with RIPA buffer. The immunoprecipitates were analyzed by immunoblotting as described above.
RESULTS
ZO-2 Down-regulates Cyclin D1 Transcription and Protein Levels
To investigate whether ZO-2 could modulate the transcription of the cyclin D1 gene, we performed reporter gene assays. For this purpose, MDCK cells were transfected with the pXP2-CD1 construct containing the human cyclin D1 regulatory region (Figure 1) driving the transcription of a luciferase reporter gene and transfected with increasing amounts of a vector expressing canine ZO-2 protein (pGW1-HA-ZO-2). Figure 2A illustrates how ZO-2 negatively affects the transcriptional activity of cyclin D1 promoter. ZO-2 specificity was tested in two ways: 1) by cotransfecting along with the pXP2-CD1 and ZO-2 plasmids a different promoter/reporter construct, pRSV-β-Gal or pSoxCAT (Figure 2B shows no significant changes in CAT or β-galactosidase reporter activity) and 2) by overexpressing ZO-1, a related member of the MAGUK family, and a member that shares with ZO-2 several structural characteristics (Gonzalez-Mariscal et al., 2000; Gonzalez-Mariscal et al., 2003) (ZO-1 overexpression did not alter promoter activity; Figure 2B).
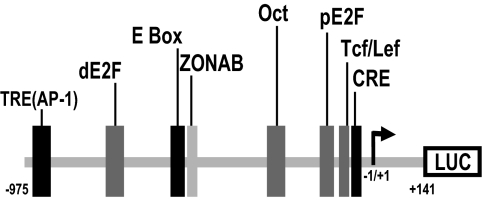
Figure 1.
Schematic representation of the human cyclin D1 promoter. The pXP2-CD1 reporter vector contains the cyclin D1 regulatory region here shown. TRE(AP-1), TRE, −924 to −938 (Herber et al., 1994); dE2F, distal E2F, −730 to −737 (Herber et al., 1994); E box, bHLH, and Zn finger transcription factors binding site, −547 to −553 (Herber et al., 1994); ZONAB, ZONAB binding site and inverted CCAAT box, −527 to −532 (Sourisseau et al., 2006); Oct, Oct binding site, −646 to −652 (Cicatiello et al., 2004); pE2F, proximal E2F, −133 to −143 (Watanabe et al., 1998); Tcf/Lef, Tcf/Lef binding site, −67 to −77 (Shtutman et al., 1999); CRE, −46 to −53, (Herber et al., 1994); and Start, site of transcription initiation, −1/+1 (Herber et al., 1994). Broken arrow stands for the transcription start point.
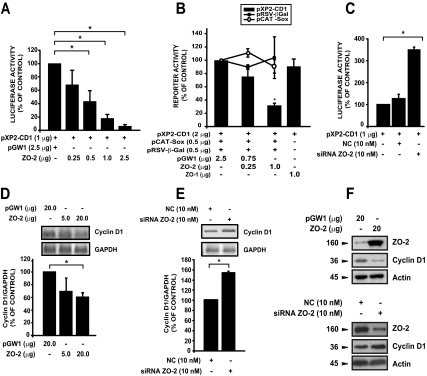
Figure 2.
ZO-2 down-regulates cyclin D1 transcription. (A) Cyclin D1 promoter activity was tested under conditions of high ZO-2 levels. MDCK cells were transiently cotransfected with 1 μg of pXP2-CD1 reporter plasmid and the indicated amounts of pGW1-HA-ZO-2 (ZO-2) expression vector or with the empty vector (pGW1). pXP2-CD1/pGW1 is taken as control. (B) To test the specificity of the ZO-2 effect, MDCK sparse monolayers were cotransfected with different reporter plasmids (pRSV-β-Gal, pCAT-Sox, or pXP2-CD1) and ZO-2 or pCB6 ZO-1myc (ZO-1) as indicated in the graph. Control reference activity was taken for each reporter as the activity under the influence of pGW1. (C) Cells were transiently transfected with 10 nM siRNA designed to target ZO-2 mRNA or a NC siRNA not homologous to anything in the vertebrate transcriptosome. Forty-eight hours after initial transfection cells were retransfected with 1 μg of pXP2-CD1 reporter plasmid. Harvesting was performed after 72 h (total time). Graphs show relative luciferase activity taken as a percentage of control (only pXP2-CD1) activity. (D and E) In vivo transcription of cyclin D1 tested in response to differential expression of ZO-2. Total RNA was extracted from monolayers transiently transfected with full-length ZO-2 (pGW1-HA-ZO-2), the empty vector (pGW1; D), or transfected with siRNA for ZO-2 or NC (E). Semiquantitative RT-PCR assays were performed to detect cyclin D1 mRNA levels. Data were quantified by densitometry of amplified bands normalizing optical densities of cyclin D1 to those from the GAPDH signal. Data shown are relative to control cells transfected only with the empty vector. (F) Cyclin D1 protein levels in MDCK cells, transfected as indicated in D and E, were monitored by Western blotting. Total protein extracts from sparse MDCK cells, transiently transfected with full-length ZO-2 or the empty vector, were analyzed by immunodetection with specific antibodies against ZO-2, cyclin D1, and actin 48 h after transfection. Numbers to the left indicate the molecular sizes in kilodaltons. Data are the mean values ± SE of at least three independent experiments; *p < 0.05, Student's t test.
To gain additional evidence of the occurrence of this phenomenon, we performed the same experiment with cells where endogenous ZO-2 had been silenced with siRNA. The ZO-2 siRNA decreased ZO-2 protein expression in MDCK cells by 65%, and this effect was apparent within 24 h and persisted for 1 week [(data not shown) (Hernandez et al., 2007)]. Figure 2C shows how ZO-2–silenced cells allowed the cyclin D1 promoter to drive a higher transcription of the luciferase reporter gene than those transfected with an NC siRNA, which was not homologous to anything in the vertebrate transcriptosome.
Concomitantly, we tested by semiquantitative RT-PCR the endogenous levels of cyclin D1 messenger to confirm that ZO-2 regulates cyclin D1 transcription. Figure 2D shows the clear reduction of cyclin D1 messenger when ZO-2 was overexpressed, whereas Figure 2E reveals how in ZO-2 knockdown cultures, there was an increase in cyclin D1 mRNA. In MDCK cells, the overexpression of ZO-2 protein also induced a diminished level of cyclin D1 protein, and opposite to this effect, siRNA of ZO-2 increased cyclin D1 protein expression (Figure 2F). Together, these data indicated that ZO-2 exerts a negative regulation of cyclin D1 transcription.
We next wondered whether changes observed in cyclin D1 reporter gene, mRNA and protein expression could have a physiological relevance on cell proliferation. To pursue this point, DNA synthesis by [3H]thymidine incorporation was determined in cells transfected with pGW1-HA-ZO-2 or the empty vector (Figure 3). In cells overexpressing ZO-2, a diminished cell proliferation rate was observed. Differences in cell proliferation rate were significant for those rates obtained between 24 and 96 h.
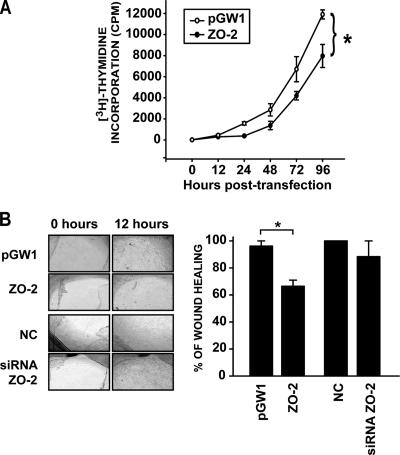
Figure 3.
Overexpression of ZO-2 modifies cell proliferation rates. (A) DNA synthesis was monitored by [3H]thymidine incorporation into MDCK cells transfected with the ZO-2 expression vector (pGW1-HA-ZO-2) or the empty vector (pGW1). [3H]Thymidine uptake was determined at various posttransfection time points. Data shown are mean ± SE values of at least three independent experiments performed in triplicate. Differences are significant beginning at 24 h. (B) Wound healing assays were performed on confluent MDCK monolayers previously transfected with 5 μg of HA-pGW1-ZO-2 or the empty vector pGW1 or with 10 nM siRNA specific for ZO-2 or the NC. Wound closure was followed by taking microscopic images at different time points after monolayer damage was inflicted. In each case, denuded areas were evaluated, quantified, and compared with obtain the percentage of wound healing (see graph). Data obtained at wound time 0 were set as control. *p < 0.05, Student's t test.
Next, we tested whether the overexpression of ZO-2 as well as the silencing of ZO-2 would affect wound closure. For this purpose, sparse MDCK cells seeded on eight-well glass slide chambers were transfected with pGW1-HA-ZO-2, the empty vector or the siRNAs for ZO-2, or the NC siRNAs unable to silence any mammalian RNA. Once these monolayers reached confluence, a wound was made with a pipette tip, and photographs of the wounded area were taken periodically. Figure 3B illustrates how the culture transfected with ZO-2 exhibited by the 12th hour a delayed healing in comparison with those transfected with the empty vector or the siRNAs. Previously, it had been shown that ZO-2 silencing exerted no effect in cell proliferation (Hernandez et al., 2007); therefore, it is not surprising that ZO-2 silencing does not alter wound healing.
ZO-2 Repression Is Mediated by a Putative E2F/E Box Located in the Distal Portion of the Cyclin D1 Promoter
Next, we proceeded to search for elements in the promoter region of cyclin D1 that could mediate the effect induced by ZO-2. Multiple transcription factors have been shown to recognize and regulate cyclin D1 gene activity (Coqueret, 2002). However, we started our study with the AP-1 binding sites TRE(AP-1) and cAMP response element (CRE), because AP-1 is an important activator of cyclin D1 transcription, and also because our previous studies had revealed the interaction of ZO-2 with Jun and Fos (AP-1 complex) and we had demonstrated inhibition of the transcriptional activity of an AP-1–dependent promoter upon ZO-2 overexpression (Betanzos et al., 2004).
Constructs with point mutations in TRE(AP-1) and CRE sites, depicted on the left side of Figure 4A, were tested under ZO-2 influence. As reported previously (Herber et al., 1994), the activity of these mutated constructs is lower than that of the wild-type promoter pXP2-CD1, indicating that indeed, activator elements were removed. With both TRE(AP-1) and CRE mutant plasmids, ZO-2 still had a negative effect over the reporter activity (Figure 4A). This result may be due to the fact that each mutant keeps one AP-1 binding site intact or could arise if ZO-2 exerts its effect by interacting with other transcription factors that bind to sites in the promoter different from TRE(AP-1) and CRE.
Inside the region located between nucleotides (nt) −737/−730 (distal) or between nt −143/−133 (proximal), there are elements recognized by the transcription factors of the E2F family (Fukami-Kobayashi and Mitsui, 1998; Watanabe et al., 1998; Cicatiello et al., 2004). Because ChIP assays (Cicatiello et al., 2004) have revealed that E2F factors bind to DNA in a cell cycle progression-related manner, we next performed deletions of the E2F region, and we designed functional assays to test the particular relevance of these transcription factors in controlling the cyclin D1 promoter activity under ZO-2 effect.
In accordance with the results found in other cell lines (Watanabe et al., 1998), the deleted distal “E2F” region seemed to contain a silencer element for the cyclin D1 promoter (Figure 4B). The generation of different constructs where the region surrounding E2F was deleted allowed the detection of a fragment located between nt −694 and −550 that is required for attaining the negative response triggered by ZO-2 (Figure 4B). In silico analysis of this region (Figure 4C) revealed the presence of an E box element that was described previously but not tested (Herber et al., 1994; Peukert et al., 1997) and of a novel putative E2F binding site. These observations suggested that ZO-2 may need an intact E box/E2F element located between nt 553 and −548 to exercise its negative effect over cyclin D1 transcription.
ZO-2 Associates and Binds In Vivo to the E Box/E2F Element in the Cyclin D1 Promoter Region
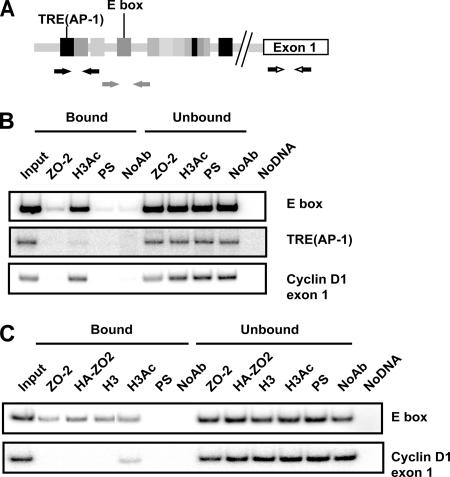
To study whether ZO-2 could associate in vivo to the TRE(AP-1) and E box/E2F elements, we performed ChIP assays amplifying the regions indicated in Figure 5A in either sparse cultures, where ZO-2 is concentrated at the nuclei (Islas et al., 2002) (Figure 5B), or in cells overexpressing ZO-2 (Figure 5C). This assay included immunoprecipitations with anti-histone 3 and acetylated histone 3 antibodies for positive controls, and immunoprecipitations with preimmune serum and without antibody as negative controls. We found that endogenous ZO-2 associates specifically to the E box element but not to the TRE(AP-1) region or the exon 1 segment of the cyclin D1 gene, which was chosen as a negative control. In a similar way, exogenous overexpressed ZO-2 associated to the E box region, demonstrating that ZO-2 binds the canine E box sequence in cyclin D1 promoter in vivo (Figure 5, B and C).
Figure 5.
ZO-2 associates in vivo to an E box element in the cyclin D1 promoter. (A) Schematic localization of specific primers used for PCR reactions in ChIP assays designed for the dog cyclin D1 promoter. Amplification of exon 1 was used as a negative control. (B) ChIP assays performed in sparse MDCK cells reveal the association of ZO-2 with an E box and not to the TRE(AP-1) element also present in the cyclin D1 promoter. The sizes of the amplified regions are 269 base pairs for the E box region, 200 base pairs for the TRE(AP-1) area, and 200 base pairs for exon 1. In this and the following ChIP assays, positive controls with H3Ac or H3 and negative controls with preimmune serum (PS) and no antibody (NoAb) were included. Input DNA, bound, unbound and no DNA fractions were amplified with the same set of primers as additional quality controls. (C) ChIP assay to detect recombinant ZO-2 were performed in cells that overexpress ZO-2 (pGW1-HA-ZO-2). Immunoprecipitates done with antibodies against ZO-2 and HA confirm the interaction of native and transfected ZO-2 (HA-tagged ZO-2) with the E box. Representative images of radioactive PCR products are shown. At least two independent experiments were performed for all ChIP assays.
These data support the idea of a physical interaction of ZO-2 with the chromatin region of the cyclin D1 promoter. Because ZO-2 does not have any DNA binding domain inside its structure, we postulated that its negative transcriptional activity might be mediated through other proteins, such as transcription factors that directly recognize DNA elements.
c-Myc Recognizes the E box/E2F Region in Cyclin D1 Promoter
The presence of the above-mentioned E box in the promoter of cyclin D1 was reported previously; yet, the importance of it as a functional element for cyclin D1 regulation had not been explored (Herber et al., 1994; Zhang et al., 2002). Transcription factors binding to E box elements include c-Myc, USF, and Snail (Blackwell et al., 1990; Nieto, 2002; Adhikary and Eilers, 2005; Corre and Galibert, 2005, 2006; De Craene et al., 2005). c-Myc forms heterodimers with Max, and both activate and repress large groups of mammalian genes by recruiting histone modifying and chromatin remodeling enzymes (for review, see Adhikary and Eilers, 2005). In fact, cyclin D1 has been found to be regulated by c-Myc in both an activating and an inhibitory manner (Adhikary and Eilers, 2005; Kleine-Kohlbrecher et al., 2006).
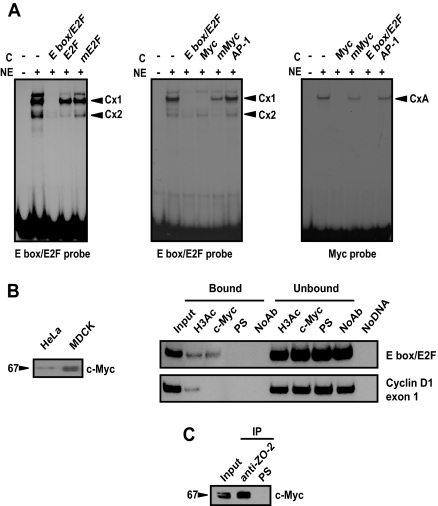
Here, we wondered whether c-Myc could bind to the E box region of the cyclin D1 promoter and whether this factor could allow ZO-2 docking. Thus, EMSAs were performed to identify the nature of the DNA–protein interactions occurring in this particular region. For this purpose, the association between an E box/E2F probe and nuclear proteins extracted from MDCK cells was tested, and the formation of two complexes (Cx1 and Cx2) was found (Figure 6A, left). The appearance of Cx2 was abolished by an excess of nonlabeled E2F consensus oligonucleotide. In Figure 6A, middle, it is possible to appreciate that the formation of Cx1 is dependent on c-Myc, because only the c-Myc consensus sequence can effectively compete for it, whereas a heterologous probe, AP-1, or the mutant c-Myc did not cause any effect. Figure 6A, right, shows a reverse competition assay that confirms binding of c-Myc to the E box/E2F region of cyclin D1 promoter.
Figure 6.
c-Myc recognizes the E box region in cyclin D1 promoter. (A) Gel shift assays done with the indicated 32P-end–labeled oligonucleotide. Left, E box/E2F probe interaction with proteins present in the nuclear extract (NE) was competed (C stands for competitor added) with a nonlabeled E box/E2F oligonucleotide and E2F consensus or mutant oligonucleotides. Middle, E box/E2F labeled oligonucleotide was competed with consensus c-Myc probe, the corresponding mutant c-Myc or AP-1 as a heterologous competitor. Right, consensus c-Myc was used as labeled probe for reverse competition. Black arrowheads on the right indicate the position of the specific complexes (Cx). All competition experiments were performed in the absence or presence of 100-fold excess of the indicated nonlabeled competitor and 10 μg of nuclear extracts obtained from sparse MDCK cells. (B) Left, 60 μg of nuclear extracts obtained from sparse MDCK and HeLa cells was subjected to Western blot using an antibody against c-Myc. A 67-kDa band was detected. Right, ChIP assay reveals the in vivo association of c-Myc with the E box present in the cyclin D1 promoter. (C) c-Myc is present in ZO-2 immunoprecipitate. Input, 10% of immunoprecipitation starting material.
To determine whether c-Myc localizes in vivo to this E box region present in the distal promoter of cyclin D1, we next performed ChIP assays. First, we demonstrated by Western blot that the c-Myc antibody used recognizes endogenous c-Myc present in nuclear extracts derived from MDCK cells (Figure 6B, left). Then, using this antibody, we performed ChIP assays and we found that c-Myc associates in vivo to the E box region (Figure 6B, middle). To further confirm the in vivo interaction established between c-Myc and ZO-2, we immunoprecipitated ZO-2 from nuclear extracts derived from MDCK cells, and we detected by Western blot the presence of c-Myc in the immunoprecipitate (Figure 6C). These experiments thus demonstrated that c-Myc binds to the E box region docking ZO-2 to the cyclin D1 promoter.
c-Myc Cooperates with ZO-2 in Down-regulating Cyclin D1 Transcription
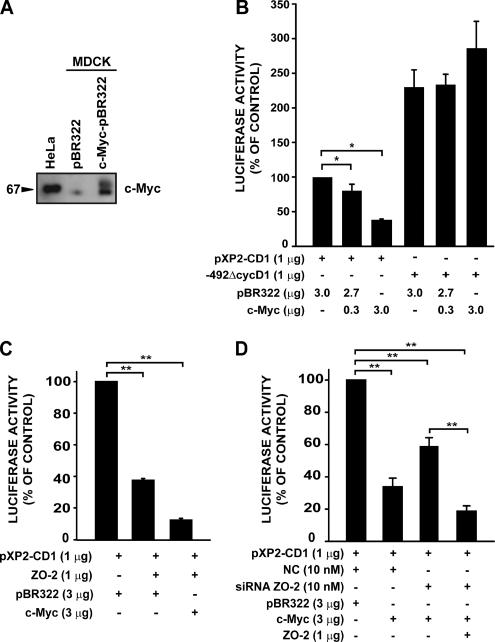
Next, we explored the effect of c-Myc on cyclin D1 promoter activity by using functional assays in MDCK cells. First, we transfected into MDCK cells a plasmid carrying human c-Myc and observed overexpression of this protein (Figure 7A). Next, we performed cotransfection assays with the pXP2-CD1 reporter and different amounts of the plasmid that overexpresses human c-Myc (Figure 7B). We found that c-Myc acts as a repressor for the cyclin D1 promoter. When we tested a deletion construct lacking the E box element, c-Myc lost its inhibitory effect on the cyclin D1 promoter. Having demonstrated that ZO-2 and c-Myc independently act as repressors for cyclin D1 transcription, we next investigated the effect of cotransfecting both proteins on cyclin D1 promoter activity.
Figure 7.
c-Myc cooperates with ZO-2 in down-regulating the cyclin D1 promoter. (A) Western blot done with c-Myc antibody reveals how c-Myc is overexpressed in MDCK cells transfected with the c-Myc-LTR/pBR322 or the empty vector LTR/pBR322 (indicated as pBR322). HeLa cell extract was included as a positive control. (B) c-Myc overexpression represses cyclin D1 promoter activity in a dose-dependent manner and displays specificity for the E box element. This assay was done with sparse MDCK cells transfected with 1 μg of the intact cyclin D1 promoter construct (pXP2-CD1) or with a plasmid lacking E box/E2F element (−492ΔCycD1) and indicated amounts of c-Myc-LTR/pBR322 plasmid or the empty vector LTR/pBR322. (C) Simultaneous overexpression of c-Myc and ZO-2 enhanced the repression of cyclin D1 promoter activity. MDCK cells were cotransfected as indicated with different combinations of the following plasmids: pXP2-CD1, ZO-2, pBR322, and c-Myc. (D) Addition of ZO-2 siRNA modifies c-Myc repression abilities. Over a constant c-Myc expression, ZO-2 siRNA but not NC interferes with repression. Recombinant ZO-2 expression abrogates the ZO-2 siRNA effect. Relative luciferase activities are expressed as a percentage of the control activity (pXP2-CD1/pBR322 or in combination with NC siRNA). Data are mean values ± SE of at least three independent experiments (*p < 0.05, **p < 0.001, Student's t test).
Figure 7C illustrates how the overexpression of both ZO-2 and c-Myc together exerted a synergic effect that enhanced the repressive effect on cyclin D1 transcription. These data thus demonstrated that c-Myc cooperates with ZO-2 in down-regulating cyclin D1 transcription.
The results shown in Figure 7D indicated that full repressor activity is achieved when c-Myc is present in combination with ZO-2. A significant recovery in activity is observed when cells were treated with ZO-2 siRNA, decreasing ZO-2 levels; overexpression of recombinant ZO-2 abolished the effect of previously transfected ZO-2 siRNA.
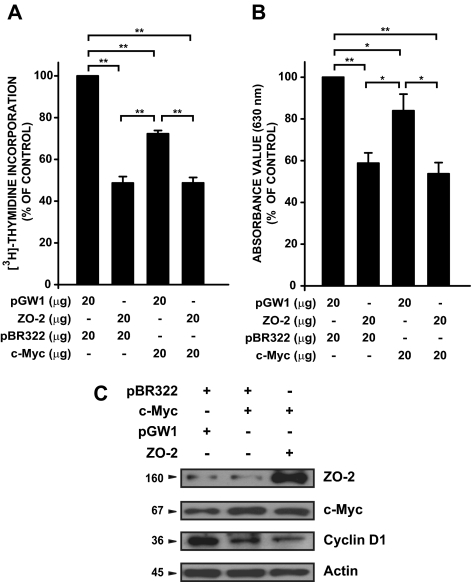
To relate c-Myc/ZO-2 cooperative repressive action on cyclin D1 with cell proliferation, MDCK cells were cotransfected with both c-Myc– and ZO-2–expressing plasmids, and cell proliferation rates were determined. [3H]Thymidine incorporation and MTT detection indicated that under this condition, cell proliferation was inhibited (Figure 8). Immunodetection of cyclin D1 indicated that coexpression of both repressors (c-Myc and ZO-2) resulted in a diminished amount of the 36-kDa protein (Figure 8C).
Figure 8.
Myc/ZO-2 down-regulates cell proliferation. Cell proliferation assessed by [3H]thymidine incorporation (A) and by MTT method (B) in MDCK cells transiently cotransfected (electroporated) with 40 μg of combined vectors for 48 h as indicated in the figure. Results are expressed as a percentage of the empty vectors value (control). Bioassays were performed with each value being determined in triplicate. Mean values ± SE of at least three independent experiments are plotted. *p < 0.05, **p < 0.001, Student's t test. (C) Immunodetection of indicated proteins was performed as described in Figure 2 in the same cells electroporated as in A or B.
HDAC1 Cooperates with ZO-2/c-Myc in Repressing Cyclin D1 Transcription
Histone acetylation and deacetylation are major regulatory transcription mechanisms that function by modulating the accessibility of transcription factors to their cognate binding sites on DNA. Consequently, a common feature of mammalian transcriptional repressors is the promoter-specific recruitment of HDACs (Ng and Bird, 2000; Free et al., 2001).
Because our results have shown that ZO-2 and c-Myc behave as repressors, we next explored whether HDAC proteins participated in this process. For this purpose, MDCK cells were cotransfected with the cyclin D1 promoter construct and the ZO-2 expression vector and then treated for 12 h with the HDAC inhibitors trichostatin A or sodium butyrate. Figure 9A shows how ZO-2 repression activity diminished in the presence of either one of the inhibitors.
Figure 9.
HDAC1 are recruited and participate in the negative effect mediated by ZO-2 and c-Myc on cyclin D1 transcription. (A) Participation of HDACs in the repression mediated by ZO-2/c-Myc. MDCK cells were transiently cotransfected with pXP2-CD1 reporter plasmid and pGW1-HA-ZO-2 and treated with HDAC inhibitors TSA (2 nM) and NaB (5 mM) for 12 h before harvesting. Results are presented as the percentage of repression exerted by ZO-2 under each treatment related to nontreated cells. Mean values ± SE of at least three independent experiments (*p < 0.05, Student's t test). (B) ChIP assay reveals the interaction of HDAC1 with the E box present in the cyclin D1 promoter. (C) HDAC1 is present in ZO-2 immunoprecipitate derived from nuclear extracts of sparse MDCK cells. A 60-kDa band was detected in the MDCK input and immunoprecipitation (IP) and in control HeLa cells. Input, 10% of immunoprecipitation starting material.
To determine whether HDAC was associated to the E box region of the cyclin D1 promoter, we performed a ChIP assay by using MDCK cells that overexpressed ZO-2. Because HDAC1 is expressed in MDCK cells (Peinado et al., 2004), we used a specific antibody directed against it. Figure 9B shows that HDAC1 is indeed recruited to the E box region when ZO-2 is overexpressed in MDCK cells. To further confirm this in vivo interaction, we performed an immunoprecipitation experiment that revealed HDAC1 was present in a ZO-2 nuclear immunoprecipitate (Figure 9C). Together, these results highlight HDAC1 as a participant in the down-regulation of cyclin D1 transcription mediated by ZO-2 and c-Myc. A model that recapitulates our findings is presented in Figure 10.
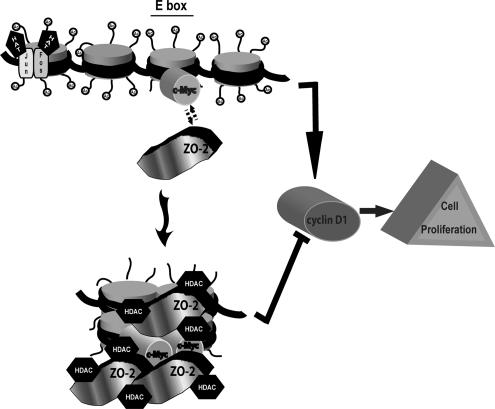
Figure 10.
ZO-2, c-Myc, and HDAC1 interact in the cyclin D1 E box promoter region blocking cell proliferation. HAT, histone acetyl transferase.
DISCUSSION
The role of ZO-2 in the nucleus has started to be unraveled. Our work with reporter genes under the control of AP-1–regulated promoters revealed the participation of ZO-2 in the repression of gene transcription (Betanzos et al., 2004). Because TJs seem to be involved in the regulation of cell proliferation, we analyzed whether ZO-2 was able to affect the transcription of cyclin D1, a key regulator of the cell cycle. Our results indicated that ZO-2 in a complex with c-Myc and HDAC1 repressed the transcription of the cyclin D1 gene. This repression seemed to be physiologically relevant, because cells overexpressing ZO-2 exhibited a diminished cell proliferation rate as determined through [3H]thymidine incorporation and wound healing assays.
The relationship among TJ proteins, gene transcription, and cell proliferation was uncovered with the pioneering work of Balda and Matter. They revealed how ZO-1 inhibited G1/S phase transition by cytoplasmic sequestration of ZONAB, a transcription factor that forms a complex with CDK4 (Balda and Matter, 2000; Balda et al., 2003). Moreover, they showed that upon ZONAB overexpression cyclin D1 gene expression became up-regulated, whereas the expression of the SH3 domain of ZO-1, which inhibits ZONAB function, induced only a small inhibition of the cyclin D1 promoter (Sourisseau et al., 2006). The latter observation is in accordance with our present findings in MDCK cells that revealed that in contrast to ZO-2, ZO-1 transfection exerted no direct effect on the activity of the human cyclin D1 promoter (Figure 2B). These results demonstrated that each of these proteins exerted a particular function not shared by other ZO proteins.
Because ZO proteins do not have the characteristic features in their sequence that allow binding to DNA, they might form complexes with other proteins, such as transcription factors, to regulate gene transcription. The interaction between ZO-1 with ZONAB and ZO-2 with Jun, Fos, and C/EBP has confirmed this proposal and has allowed the exploration of regulatory regions of genes regulated by ZO protein expression.
Our work initially explored TRE(AP-1) and CRE sites of the human cyclin D1 promoter, located at positions −929 to −935 and −45 to −52, respectively. Because the mutation of just one of any of these sites generated a profound loss of promoter activity, it became extremely difficult to appreciate the impact ZO-2 overexpression had on them (Figure 4A). However, we further pursued this point in a ChIP assay done by immunoprecipitating ZO-2 and performing PCR amplification with primers for the TRE site. Because no amplification was obtained, we suspect that ZO-2 is not exerting its inhibitory effect on cyclin D1 transcription through association to the TRE(AP-1) site (Figure 5B). With regard to the CRE site, it should be pointed out that the construct containing the deletion of the E box and that showed no repression upon ZO-2 overexpression still has an intact CRE site (Figure 4B). These results, therefore, suggested that this location is not involved in the inhibition of cyclin D1 transcription triggered by ZO-2 overexpression.
The deletion analysis performed around the E2F site highlighted the importance of the E box in the transcriptional regulation of cyclin D1 modulated by ZO-2 (Figure 4). Structurally different factors of the basic-helix loop-helix (bHLH) type, like c-Myc and USF, or the zinc finger protein family, like Snail, recognize the CAC(A/G)TG sequence that characterizes E box elements (Blackwell et al., 1990; Cole and McMahon, 1999; Nieto, 2002; Corre and Galibert, 2005, 2006). We discarded the participation of Snail in the repression of cyclin D1 mediated by ZO-2, because Snail protein levels are so low in MDCK cells that they escape detection with commercial antibodies (Comijn et al., 2001; Bolos et al., 2003). However, to further address this issue, we performed ChIP assays in MDCK cells that overexpress Snail (generously provided by professor Roberto Montesano, Department of Cell Physiology and Metabolism, University of Geneva Medical Center), and we did not observe interaction of Snail with the E box region of the cyclin D1 promoter (data not shown). The participation of USF and/or E2F transcription factors in cyclin D1 down-regulation by ZO-2 is a matter of current work in our laboratory, because at least in the case of E2F, the electrophoretic mobility shift assays showed effective competition of this factor for complex 2 (Figure 6A, left).
With regards to c-Myc, we demonstrated its ability to bind to the E box element of the cyclin D1 promoter, to repress the promoter activity either alone or in a cooperative manner with ZO-2 (Figures 6 and 7), and to inhibit cell proliferation (Figure 8). Another tumor suppressor known to modulate the transcriptional activity regulated by c-Myc is, for example, Bin1, which interacts with the N-terminal region of c-Myc and inhibits malignant cell proliferation (Elliott et al., 1999).
Besides cyclin D1 (Philipp et al., 1994), other genes repressed in c-Myc–transformed cells included those that encode cell adhesion proteins, such as the β1 integrin subunit (Judware and Culp, 1995); molecules of the immune system, such as major histocompatibility complex class I and lymphocyte function-associated antigen-1 (Bernards et al., 1986; Versteeg et al., 1988; Inghirami et al., 1990); and cell cycle regulators, such as C/EBPα (Freytag and Geddes, 1992) and c-Myc itself (Penn et al., 1990). These findings suggested that gene repression contributes significantly to the phenotype of c-Myc transformed cells. Formation of Myc:Max heterodimers is essential for both Myc-dependent transcriptional activation and gene repression (Mao et al., 2003). Other combinations of c-Myc heterodimers have been described that recruit remodeling enzymes, such as HDACs, and exert a repressing effect. Such is the case of MM-1, a novel c-Myc-binding protein that interferes with the E box-dependent transcription activity of c-Myc (Mori et al., 1998). In HeLa cells, c-Myc repression is controlled by a complex containing c-Myc, MM-1, the corepressors mSin3 and TIF1β, and HDAC1 (Satou et al., 2001). Here, we have described the formation of a complex consisting of ZO-2, c-Myc, and HDAC1 that participates in the negative regulation of cyclin D1; however, we cannot rule out the inclusion of other components in this complex.
In summary, we can conclude that ZO-2 inhibits the transcription of cyclin D1 by forming a complex that includes c-Myc and HDAC1, and this complex associates with the E box present in the cyclin D1 promoter (Figure 10). The interaction of ZO-2 with this complex is important for understanding the role played by TJ proteins in the regulation of cell proliferation and tumorigenesis.
ACKNOWLEDGMENTS
We acknowledge the technical assistance of Matilde Corona, Gerardo Marmolejo, Socorro Islas, Guadalupe Aguilar, Ricardo Valle, Georgina Guerrero, Inti de la Rosa, and Hector Rincón. Miriam Huerta, Rodrigo Muñoz, Rocío Tapia, and Ernesto Soto-Reyes were recipients of doctoral fellowships from the Consejo Nacional de Ciencia y Tecnologia (CONACyT) (159492, 183731, 166727, and 181389). We thank Irma Cruz-Solis for the kind donation of pSoxCAT. This work was supported by grants 41273-A and 50414 from CONACyT (to E.L.-B.) and 45691-Q (to L.G.-M.).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-02-0109) on September 19, 2007.
REFERENCES
- Adhikary S., Eilers M. Transcriptional regulation and transformation by Myc proteins. Nat. Rev. Mol. Cell Biol. 2005;6:635–645. doi: 10.1038/nrm1703. [DOI] [PubMed] [Google Scholar]
- Albanese C., et al. Activation of the cyclin D1 gene by the E1A-associated protein p300 through AP-1 inhibits cellular apoptosis. J. Biol. Chem. 1999;274:34186–34195. doi: 10.1074/jbc.274.48.34186. [DOI] [PubMed] [Google Scholar]
- Albanese C., Johnson J., Watanabe G., Eklund N., Vu D., Arnold A., Pestell R. G. Transforming p21ras mutants and c-Ets-2 activate the cyclin D1 promoter through distinguishable regions. J. Biol. Chem. 1995;270:23589–23597. doi: 10.1074/jbc.270.40.23589. [DOI] [PubMed] [Google Scholar]
- Arber N., Hibshoosh H., Moss S. F., Sutter T., Zhang Y., Begg M., Wang S., Weinstein I. B., Holt P. R. Increased expression of cyclin D1 is an early event in multistage colorectal carcinogenesis. Gastroenterology. 1996;110:669–674. doi: 10.1053/gast.1996.v110.pm8608874. [DOI] [PubMed] [Google Scholar]
- Balda M. S., Garrett M. D., Matter K. The ZO-1-associated Y-box factor ZONAB regulates epithelial cell proliferation and cell density. J. Cell Biol. 2003;160:423–432. doi: 10.1083/jcb.200210020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balda M. S., Matter K. The tight junction protein ZO-1 and an interacting transcription factor regulate ErbB-2 expression. EMBO J. 2000;19:2024–2033. doi: 10.1093/emboj/19.9.2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernards R., Dessain S. K., Weinberg R. A. N-myc amplification causes down-modulation of MHC class I antigen expression in neuroblastoma. Cell. 1986;47:667–674. doi: 10.1016/0092-8674(86)90509-x. [DOI] [PubMed] [Google Scholar]
- Betanzos A., Huerta M., Lopez-Bayghen E., Azuara E., Amerena J., Gonzalez-Mariscal L. The tight junction protein ZO-2 associates with Jun, Fos and C/EBP transcription factors in epithelial cells. Exp. Cell Res. 2004;292:51–66. doi: 10.1016/j.yexcr.2003.08.007. [DOI] [PubMed] [Google Scholar]
- Blackwell T. K., Kretzner L., Blackwood E. M., Eisenman R. N., Weintraub H. Sequence-specific DNA binding by the c-Myc protein. Science. 1990;250:1149–1151. doi: 10.1126/science.2251503. [DOI] [PubMed] [Google Scholar]
- Bolos V., Peinado H., Perez-Moreno M. A., Fraga M. F., Esteller M., Cano A. The transcription factor Slug represses E-cadherin expression and induces epithelial to mesenchymal transitions: a comparison with Snail and E47 repressors. J. Cell Sci. 2003;116:499–511. doi: 10.1242/jcs.00224. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Cicatiello L., et al. Estrogens and progesterone promote persistent CCND1 gene activation during G1 by inducing transcriptional derepression via c-Jun/c-Fos/estrogen receptor (progesterone receptor) complex assembly to a distal regulatory element and recruitment of cyclin D1 to its own gene promoter. Mol. Cell. Biol. 2004;24:7260–7274. doi: 10.1128/MCB.24.16.7260-7274.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole M. D., McMahon S. B. The Myc oncoprotein: a critical evaluation of transactivation and target gene regulation. Oncogene. 1999;18:2916–2924. doi: 10.1038/sj.onc.1202748. [DOI] [PubMed] [Google Scholar]
- Comijn J., Berx G., Vermassen P., Verschueren K., van Grunsven L., Bruyneel E., Mareel M., Huylebroeck D., van Roy F. The two-handed E box binding zinc finger protein SIP1 downregulates E-cadherin and induces invasion. Mol. Cell. 2001;7:1267–1278. doi: 10.1016/s1097-2765(01)00260-x. [DOI] [PubMed] [Google Scholar]
- Coqueret O. Linking cyclins to transcriptional control. Gene. 2002;299:35–55. doi: 10.1016/s0378-1119(02)01055-7. [DOI] [PubMed] [Google Scholar]
- Corre S., Galibert M. D. Upstream stimulating factors: highly versatile stress-responsive transcription factors. Pigment Cell Res. 2005;18:337–348. doi: 10.1111/j.1600-0749.2005.00262.x. [DOI] [PubMed] [Google Scholar]
- Corre S., Galibert M. D. USF as a key regulatory element of gene expression. Med. Sci. 2006;22:62–67. doi: 10.1051/medsci/200622162. [DOI] [PubMed] [Google Scholar]
- Chlenski A., Ketels K. V., Engeriser J. L., Talamonti M. S., Tsao M. S., Koutnikova H., Oyasu R., Scarpelli D. G. zo-2 gene alternative promoters in normal and neoplastic human pancreatic duct cells. Int. J. Cancer. 1999;83:349–358. doi: 10.1002/(sici)1097-0215(19991029)83:3<349::aid-ijc10>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- Chlenski A., Ketels K. V., Korovaitseva G. I., Talamonti M. S., Oyasu R., Scarpelli D. G. Organization and expression of the human zo-2 gene (tjp-2) in normal and neoplastic tissues. Biochim. Biophys. Acta. 2000;1493:319–324. doi: 10.1016/s0167-4781(00)00185-8. [DOI] [PubMed] [Google Scholar]
- De Craene B., van Roy F., Berx G. Unraveling signalling cascades for the Snail family of transcription factors. Cell Signal. 2005;17:535–547. doi: 10.1016/j.cellsig.2004.10.011. [DOI] [PubMed] [Google Scholar]
- Elliott K., et al. Bin1 functionally interacts with Myc and inhibits cell proliferation via multiple mechanisms. Oncogene. 1999;18:3564–3573. doi: 10.1038/sj.onc.1202670. [DOI] [PubMed] [Google Scholar]
- Free A., Grunstein M., Bird A., Vogelauer M. Histone deacetylation: mechanisms of repression. In: Elgin S.R.C., Workman J. L., editors. Chromatin Structure and Gene Expression. New York: Oxford University Press; 2001. pp. 156–181. [Google Scholar]
- Freytag S. O., Geddes T. J. Reciprocal regulation of adipogenesis by Myc and C/EBP alpha. Science. 1992;256:379–382. doi: 10.1126/science.256.5055.379. [DOI] [PubMed] [Google Scholar]
- Fukami-Kobayashi J., Mitsui Y. The regulation of cyclin D1 expression in senescent human fibroblasts. Exp. Cell Res. 1998;241:435–444. doi: 10.1006/excr.1998.4079. [DOI] [PubMed] [Google Scholar]
- Glaunsinger B. A., Weiss R. S., Lee S. S., Javier R. Link of the unique oncogenic properties of adenovirus type 9 E4-ORF1 to a select interaction with the candidate tumor suppressor protein ZO-2. EMBO J. 2001;20:5578–5586. doi: 10.1093/emboj/20.20.5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Mariscal L., Betanzos A., Avila-Flores A. MAGUK proteins: structure and role in the tight junction. Semin. Cell Dev. Biol. 2000;11:315–324. doi: 10.1006/scdb.2000.0178. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Mariscal L., Betanzos A., Nava P., Jaramillo B. E. Tight junction proteins. Prog. Biophys. Mol. Biol. 2003;81:1–44. doi: 10.1016/s0079-6107(02)00037-8. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Mariscal L., Ponce A., Alarcon L., Jaramillo B. E. The tight junction protein ZO-2 has several functional nuclear export signals. Exp. Cell Res. 2006;312:3323–3335. doi: 10.1016/j.yexcr.2006.07.006. [DOI] [PubMed] [Google Scholar]
- Harbour J. W., Dean D. C. The Rb/E2F pathway: expanding roles and emerging paradigms. Genes Dev. 2000;14:2393–2409. doi: 10.1101/gad.813200. [DOI] [PubMed] [Google Scholar]
- Herber B., Truss M., Beato M., Muller R. Inducible regulatory elements in the human cyclin D1 promoter. Oncogene. 1994;9:2105–2107. [PubMed] [Google Scholar]
- Hernandez S., Chavez Munguia B., Gonzalez-Mariscal L. ZO-2 silencing in epithelial cells perturbs the gate and fence function of tight junctions and leads to an atypical monolayer architecture. Exp. Cell Res. 2007;313:1533–1547. doi: 10.1016/j.yexcr.2007.01.026. [DOI] [PubMed] [Google Scholar]
- Hsu E. M., McNicol P. J., Guijon F. B., Paraskevas M. Quantification of HPV-16 E6-E7 transcription in cervical intraepithelial neoplasia by reverse transcriptase polymerase chain reaction. Int. J. Cancer. 1993;55:397–401. doi: 10.1002/ijc.2910550311. [DOI] [PubMed] [Google Scholar]
- Inghirami G., Grignani F., Sternas L., Lombardi L., Knowles D. M., Dalla-Favera R. Down-regulation of LFA-1 adhesion receptors by C-myc oncogene in human B lymphoblastoid cells. Science. 1990;250:682–686. doi: 10.1126/science.2237417. [DOI] [PubMed] [Google Scholar]
- Islas S., Vega J., Ponce L., Gonzalez-Mariscal L. Nuclear localization of the tight junction protein ZO-2 in epithelial cells. Exp. Cell Res. 2002;274:138–148. doi: 10.1006/excr.2001.5457. [DOI] [PubMed] [Google Scholar]
- Jaramillo B. E., Ponce A., Moreno J., Betanzos A., Huerta M., Lopez-Bayghen E., Gonzalez-Mariscal L. Characterization of the tight junction protein ZO-2 localized at the nucleus of epithelial cells. Exp. Cell Res. 2004;297:247–258. doi: 10.1016/j.yexcr.2004.03.021. [DOI] [PubMed] [Google Scholar]
- Jones T. R., Cole M. D. Rapid cytoplasmic turnover of c-myc mRNA: requirement of the 3′ untranslated sequences. Mol. Cell. Biol. 1987;7:4513–4521. doi: 10.1128/mcb.7.12.4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judware R., Culp L. A. Over-expression of transfected N-myc oncogene in human SKNSH neuroblastoma cells down-regulates expression of beta integrin subunit. Oncogene. 1995;11:2599–2607. [PubMed] [Google Scholar]
- Kleine-Kohlbrecher D., Adhikary S., Eilers M. Mechanisms of transcriptional repression by Myc. Curr. Top. Microbiol. Immunol. 2006;302:51–62. doi: 10.1007/3-540-32952-8_3. [DOI] [PubMed] [Google Scholar]
- Lopez-Bayghen E., Vega A., Cadena A., Granados S. E., Jave L. F., Gariglio P., Alvarez-Salas L. M. Transcriptional analysis of the 5′-noncoding region of the human involucrin gene. J. Biol. Chem. 1996;271:512–520. doi: 10.1074/jbc.271.1.512. [DOI] [PubMed] [Google Scholar]
- Mao D. Y., Watson J. D., Yan P. S., Barsyte-Lovejoy D., Khosravi F., Wong W. W., Farnham P. J., Huang T. H., Penn L. Z. Analysis of Myc bound loci identified by CpG island arrays shows that Max is essential for Myc-dependent repression. Curr. Biol. 2003;13:882–886. doi: 10.1016/s0960-9822(03)00297-5. [DOI] [PubMed] [Google Scholar]
- Mori K., Maeda Y., Kitaura H., Taira T., Iguchi-Ariga S. M., Ariga H. MM-1, a novel c-Myc-associating protein that represses transcriptional activity of c-Myc. J. Biol. Chem. 1998;273:29794–29800. doi: 10.1074/jbc.273.45.29794. [DOI] [PubMed] [Google Scholar]
- Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J. Immunol. Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- Ng H. H., Bird A. Histone deacetylases: silencers for hire. Trends Biochem. Sci. 2000;25:121–126. doi: 10.1016/s0968-0004(00)01551-6. [DOI] [PubMed] [Google Scholar]
- Nieto M. A. The snail superfamily of zinc-finger transcription factors. Nat. Rev. Mol. Cell Biol. 2002;3:155–166. doi: 10.1038/nrm757. [DOI] [PubMed] [Google Scholar]
- Ohtsubo M., Roberts J. M. Cyclin-dependent regulation of G1 in mammalian fibroblasts. Science. 1993;259:1908–1912. doi: 10.1126/science.8384376. [DOI] [PubMed] [Google Scholar]
- Peinado H., Ballestar E., Esteller M., Cano A. Snail mediates E-cadherin repression by the recruitment of the Sin3A/histone deacetylase 1 (HDAC1)/HDAC2 complex. Mol. Cell. Biol. 2004;24:306–319. doi: 10.1128/MCB.24.1.306-319.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn L. J., Brooks M. W., Laufer E. M., Land H. Negative autoregulation of c-myc transcription. EMBO J. 1990;9:1113–1121. doi: 10.1002/j.1460-2075.1990.tb08217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peukert K., Staller P., Schneider A., Carmichael G., Hanel F., Eilers M. An alternative pathway for gene regulation by Myc. EMBO J. 1997;16:5672–5686. doi: 10.1093/emboj/16.18.5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philipp A., Schneider A., Vasrik I., Finke K., Xiong Y., Beach D., Alitalo K., Eilers M. Repression of cyclin D 1, a novel function of MYC. Mol. Cell. Biol. 1994;14:4032–4043. doi: 10.1128/mcb.14.6.4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quelle D. E., Ashmun R. A., Shurtleff S. A., Kato J. Y., Bar-Sagi D., Roussel M. F., Sherr C. J. Overexpression of mouse D-type cyclins accelerates G1 phase in rodent fibroblasts. Genes Dev. 1993;7:1559–1571. doi: 10.1101/gad.7.8.1559. [DOI] [PubMed] [Google Scholar]
- Satou A., Taira T., Iguchi-Ariga S. M., Ariga H. A novel transrepression pathway of c-Myc. Recruitment of a transcriptional corepressor complex to c-Myc by MM-1, a c-Myc-binding protein. J. Biol. Chem. 2001;276:46562–46567. doi: 10.1074/jbc.M104937200. [DOI] [PubMed] [Google Scholar]
- Shtutman M., Zhurinsky J., Simcha I., Albanese C., D'Amico M., Pestell R., Ben-Ze'ev A. The cyclin D1 gene is a target of the beta-catenin/LEF-1 pathway. Proc. Natl. Acad. Sci. USA. 1999;96:5522–5527. doi: 10.1073/pnas.96.10.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sourisseau T., Georgiadis A., Tsapara A., Ali R. R., Pestell R., Matter K., Balda M. S. Regulation of PCNA and cyclin D1 expression and epithelial morphogenesis by the ZO-1-regulated transcription factor ZONAB/DbpA. Mol. Cell. Biol. 2006;26:2387–2398. doi: 10.1128/MCB.26.6.2387-2398.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thierry F., Spyrou G., Yaniv M., Howley P. Two AP1 sites binding JunB are essential for human papillomavirus type 18 transcription in keratinocytes. J. Virol. 1992;66:3740–3748. doi: 10.1128/jvi.66.6.3740-3748.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traweger A., Fang D., Liu Y. C., Stelzhammer W., Krizbai I. A., Fresser F., Bauer H. C., Bauer H. The tight junction-specific protein occludin is a functional target of the E3 ubiquitin-protein ligase itch. J. Biol. Chem. 2002;277:10201–10208. doi: 10.1074/jbc.M111384200. [DOI] [PubMed] [Google Scholar]
- Versteeg R., Noordermeer I. A., Kruse-Wolters M., Ruiter D. J., Schrier P. I. c-myc down-regulates class I HLA expression in human melanomas. EMBO J. 1988;7:1023–1029. doi: 10.1002/j.1460-2075.1988.tb02909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. D., Shi S. T., Zhou Q., Goldstein S., Hong J. Y., Shao P., Qiu S. L., Yang C. S. Changes in p53 and cyclin D1 protein levels and cell proliferation in different stages of human esophageal and gastric-cardia carcinogenesis. Int. J Cancer. 1994;59:514–519. doi: 10.1002/ijc.2910590414. [DOI] [PubMed] [Google Scholar]
- Watanabe G., Albanese C., Lee R. J., Reutens A., Vairo G., Henglein B., Pestell R. G. Inhibition of cyclin D1 kinase activity is associated with E2F-mediated inhibition of cyclin D1 promoter activity through E2F and Sp1. Mol. Cell. Biol. 1998;18:3212–3222. doi: 10.1128/mcb.18.6.3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe G., Howe A., Lee R. J., Albanese C., Shu I. W., Karnezis A. N., Zon L., Kyriakis J., Rundell K., Pestell R. G. Induction of cyclin D1 by simian virus 40 small tumor antigen. Proc. Natl. Acad. Sci. USA. 1996;93:12861–12866. doi: 10.1073/pnas.93.23.12861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Q., Miskimins W. K., Miskimins R. Sox10 acts as a tissue-specific transcription factor enhancing activation of the myelin basic protein gene promoter by p27Kip1 and Sp1. J. Neurosci. Res. 2004;78:796–802. doi: 10.1002/jnr.20342. [DOI] [PubMed] [Google Scholar]
- Willott E., Balda M. S., Fanning A. S., Jameson B., Van Itallie C., Anderson J. M. The tight junction protein ZO-1 is homologous to the Drosophila discs-large tumor suppressor protein of septate junctions. Proc. Natl. Acad. Sci. USA. 1993;90:7834–7838. doi: 10.1073/pnas.90.16.7834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S. Y., Liu S. C., Goodrow T., Morris R., Klein-Szanto A. J. Increased expression of G1 cyclins and cyclin-dependent kinases during tumor progression of chemically induced mouse skin neoplasms. Mol. Carcinog. 1997;18:142–152. [PubMed] [Google Scholar]
- Zhang Z. K., Davies K. P., Allen J., Zhu L., Pestell R. G., Zagzag D., Kalpana G. V. Cell cycle arrest and repression of cyclin D1 transcription by INI1/hSNF5. Mol. Cell. Biol. 2002;22:5975–5988. doi: 10.1128/MCB.22.16.5975-5988.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]