Abstract
The INK4a-ARF locus encodes two proteins, p16INK4a and p19ARF, that restrain cell growth by affecting the functions of the retinoblastoma protein and p53, respectively. Disruption of this locus by deletions or point mutations is a common event in human cancer, perhaps second only to the loss of p53. Using insect cells infected with baculovirus vectors and NIH 3T3 fibroblasts infected with ARF retrovirus, we determined that mouse p19ARF can interact directly with p53, as well as with the p53 regulator mdm2. ARF can bind p53-DNA complexes, and it depends upon functional p53 to transcriptionally induce mdm2 and the cyclin-dependent kinase inhibitor p21Cip1, and to arrest cell proliferation. Binding of p19ARF to p53 requires the ARF N-terminal domain (amino acids 1–62) that is necessary and sufficient to induce cell cycle arrest. Overexpression of p19ARF in wild type or ARF-null mouse embryo fibroblasts increases the half-life of p53 from 15 to ≈75 min, correlating with an increased p53-dependent transcriptional response and growth arrest. Surprisingly, when overexpressed at supra-physiologic levels after introduction into ARF-null NIH 3T3 cells or mouse embryo fibroblasts, the p53 protein is handicapped in inducing this checkpoint response. In this setting, reintroduction of p19ARF restores p53’s ability to induce p21Cip1 and mdm2, implying that, in addition to stabilizing p53, ARF modulates p53-dependent function through an additional mechanism.
The INK4a-ARF locus encodes two unrelated tumor suppressor proteins, p16INK4a (1) and p19ARF (2) that act to modify the activities of the retinoblastoma protein and p53, respectively. Whereas the p53 gene is mutated in ≈50% of human cancers (3), disruption of one or more elements within the INK4a-ARF locus occurs almost as frequently in an equally broad range of tumor types (4). INK4a-ARF contains two promoters and alternative first exons, designated 1α and 1β, whose RNA products are each spliced to two common exons (2, 5–7). Exon 1α, 2, and 3 encode p16INK4a, a protein that specifically inhibits the ability of cyclin D-dependent kinases (1) to phosphorylate retinoblastoma protein (8–10). Increased expression of p16INK4a can arrest cells in the G1 phase of the cell cycle, but cells that lack functional retinoblastoma protein are resistant to p16’s effects (11–13). In contrast, the exon 1β-2–3 transcript encodes p19ARF, which bears no homology to p16INK4a and is composed of a 64-amino acid N-terminal domain derived from exon-1β and 105 C-terminal amino acids encoded by the alternative reading frame of exon 2. The p19ARF protein can induce both G1 and G2 phase arrest (2) in a manner that depends on functional p53 (14). However, ARF-null cells exhibit an intact p53-dependent G1 checkpoint in response to DNA damage by ionizing radiation (14), so ARF must lie on a different signaling pathway.
While neither p16INK4a nor p19ARF are detectably expressed during mouse embryogenesis, explantation of mouse embryo fibroblasts (MEFs) into culture induces the synthesis of both proteins (14, 15). They accumulate progressively as MEFs are passaged, during which time cell proliferation slows and eventually stops. Rare cells that spontaneously bypass this block give rise to established cell lines (16). In ≈75% of cases, establishment of cell lines is associated with loss of p53 function (17), while many of the remaining fraction sustain biallelic loss of the INK4a-ARF locus (14, 15). Although there was reason to infer that establishment of MEF-derived cell lines resulted from p16INK4a loss (18), surprisingly, deletion of ARF alone is sufficient to enable MEFs to grow, despite continued p16INK4a expression (14). In MEF strains, then, ARF and p53 can act epistatically to govern the number of allotted population doublings, with loss of either facilitating establishment. Interestingly, MEFs that lack p53 function rapidly become polyploid (19–23), but those that delete ARF tend to remain pseudodiploid (14, 15), implying that loss of p53 contributes separately to genetic instability. In agreement, tumors arising spontaneously in ARF-null mice can subsequently sustain p53 loss, indicating that p19ARF and p53 can collaborate in multistep carcinogenesis (14).
Recently, two groups reported that ARF can bind to mdm2, establishing the first direct biochemical connection between ARF and p53 (24, 25). Mdm2 (or hdm2 in humans) is encoded by a p53-responsive gene and acts in a feedback loop (26) to limit p53 function by inhibiting its transcriptional activity (27–29) and triggering its degradation (30, 31). ARF can stabilize p53 by antagonizing mdm2’s effects (24, 25). Here, we show that p19ARF can directly associate with p53 as well as with mdm2 and suggest that ARF regulates cellular functions other than p53 stabilization.
MATERIALS AND METHODS
Cell Culture.
Mammalian cells were maintained in DMEM plus 10% fetal bovine serum, 2 mM glutamine, and 100 units/ml penicillin and streptomycin (GIBCO). Balb-3T3 (10)1 cells (ARF wild-type, p53-deleted) were a gift of Arnold Levine (Princeton University, Princeton, NJ). MEFs at passages 6–9 were established as described (14); those lacking p53 came from embryos of p53-null mice (The Jackson Laboratory). Charles Sawyers (University of California, Los Angeles) provided helper and vector retrovirus plasmids. Virus production and infection were performed as previously described, and growth arrest was determined by measurements of DNA content or [3H]-thymidine incorporation into replicating DNA 48 hr postinfection (2, 14).
Spodoptera frugiperda Sf9 cells were maintained in Grace’s medium supplemented with 5% fetal bovine serum and infected for 48 hr with the indicated baculoviruses before lysis (8). Baculoviruses encoding mutant forms of p53 included one (D281G) that is defective in DNA binding and another (L22Q, W23S) that cannot bind to mdm2 (32, 33). HA-tagged ARF cDNA and deletion mutants containing (N62) or lacking (Δ1–62) the N-terminal 62 amino acids (34) were transferred from mammalian to baculovirus expression vectors.
Kinetics of p53 turnover.
MEFs were metabolically labeled for 1 hr with 200 μCi/ml [35S]methionine (1,369 Ci/mmol, ICN; 1 Ci = 37 GBq), washed and refed with DMEM containing 10% fetal bovine serum and 2 mM unlabeled methionine. Lysates of radiolabeled cells were immunoprecipitated, and recovered proteins were electrophoretically resolved in denaturing polyacrylamide gels (14).
Transactivation Assay.
NIH 3T3 cells or Balb-3T3 (10)1 cells (5 × 105 cells/100-mm diameter plate) were transfected (35) with cytomegalovirus-p53 (36) or pSRαMSV-ARF/TK-CD8 (14) vectors plus 1 μg reporter plasmid encoding chloramphenicol acetyltransferase (CAT) (37). The reporter (PG13) contains 13 repeats of a p53-specific DNA binding site in its promoter, whereas a mutant (MG15) contains 15 altered sites unable to confer p53 binding (37). Carrier plasmid was used to adjust DNA concentrations to 20 μg per plate. Cells were lysed in 0.25 M Tris⋅HCl (pH 8) by three cycles of freezing (−80°C) and thawing (25°C) 24 hr after transfection. Equal quantities of protein, determined by the Bradford method (Bio-Rad), were assayed for CAT activity by using 0.1 μCi 14C-chloramphenicol and 4 mM acetyl CoA (38), and separated products were detected by autoradiography (39).
Electrophoretic Mobility Shift Assay (EMSA).
EMSA was performed (40) with a synthetic double-stranded oligonucleotide (5′-AGGCATGCCTAGGCATGCCT) containing two p53 consensus binding sites, end-labeled with [γ32P]ATP using T4 polynucleotide kinase. Affinity-purified p53 (100 ng) (41) was mixed with lysate (1 μg protein in 5 μl EMSA buffer) from infected Sf9 cells and incubated at 25°C for 15 min in the presence or absence of the p53-activating monoclonal antibody, PAb421. Excess [32P]labeled probe (2.5 × 105 dpm in 1 μl) was added for 15 min with EMSA buffer [20 mM Hepes, pH 7.9/25 mM KCl/0.1 mM EDTA, pH 8/2 mM MgCl2/0.5 mM DTT/0.25% Nonidet P-40/10% glycerol/0.1 ng BSA/60 ng polyd(I-C)] and adjusted to a volume of 20 μl. For competition, unlabeled probe was added in 10-fold molar excess over labeled probe. Reaction mixtures (with 0.05% bromphenol blue) were loaded onto native 4% polyacrylamide gels in 45 mM Tris⋅HCl, 45 mM Na borate, 1 mM EDTA, 0.05% Nonidet P-40, and separated by electrophoresis at 250 V for 5 hr in the same buffer.
Protein Expression and Binding.
Cell pellets were lysed on ice in Tween-20 lysis buffer (50 mM Hepes, pH 7.5/150 mM NaCl/1 mM EDTA/2.5 mM EGTA/0.1% Tween-20/1 mM phenylmethylsulfonyl fluoride/0.4 units/ml aprotinin/10 μg/ml pepstatin/10 μg/ml leupeptin) and sonicated 2 × 7 sec (VirTis VirSonic 475, 12–14% power). Nuclei and debris were removed by sedimentation at 4°C in a microcentrifuge (2 min at 15,000 rpm), and protein was quantified as above. Samples (200 μg protein) electrophoretically separated on denaturing polyacrylamide gels containing SDS were transferred to Immobilon polyvinylidene difluoride membranes (Millipore) preactivated for 15 sec in methanol. Filters were washed in TBS-Tween (10 mM Tris⋅HCl, pH 7.4/150 mM NaCl/0.1% Tween-20) and blocked in the same solution containing 10% (wt/vol) nonfat dry milk. Filters exposed for 1 hr at room temperature to 0.2 μg/ml affinity-purified rabbit antibody to the mouse p19ARF C terminus (2) in TBS-Tween were washed for 45 min in TBS-Tween and incubated for 45 min with a 1/2,000 dilution of donkey antibodies to rabbit IgG (Amersham) in TBS-Tween containing 5% milk. Filters were washed and antibody binding sites were visualized by enhanced chemiluminescence as per the manufacturer’s instructions (Amersham).
For analysis of p53, mdm2, and p21Cip1 expression, frozen mammalian cell pellets were disrupted in ice-cold Nonidet P-40 lysis buffer (50 mM Tris⋅HCl, pH 8/5 mM EDTA/150 mM NaCl/0.5% Nonidet P-40/1 mM phenylmethylsulfonyl fluoride/0.4 units/ml aprotinin/10 mM β-glycerophosphate/1 mM Na/0.1 mM NaVO4), and left for 1 hr on ice. Sf9 cells were lysed by scraping in cold Tween-20 lysis buffer and sonication. Centrifuged lysates were incubated for 2 hr at 4°C with antibodies against p53 (PAb421, Calbiochem), mdm2 (monoclonal 2A10), or p21Cip1 (F5, Santa Cruz Biotechnology), respectively, plus 40 mg/ml BSA. Complexes precipitated with protein A-Sepharose were washed three times with ice-cold Nonidet P-40 lysis buffer (for mammalian lysates) or RIPA buffer (50 mM Tris⋅HCl, pH 8/150 mM NaCl/1% Triton/0.5% sodium deoxycholate/0.1% SDS/1 mM phenylmethylsulfonyl fluoride/0.4 units/ml aprotinin/10 μg/ml pepstatin/10 μg/ml leupeptin) (for Sf9 lysates). Precipitates were separated on denaturing polyacrylamide gels and transferred to nitrocellulose. Mdm2 and p21Cip1 were detected by immunoblotting with the same antibodies, and p53 with Ab-7 (Calbiochem), visualized by chemiluminescence as above.
RESULTS
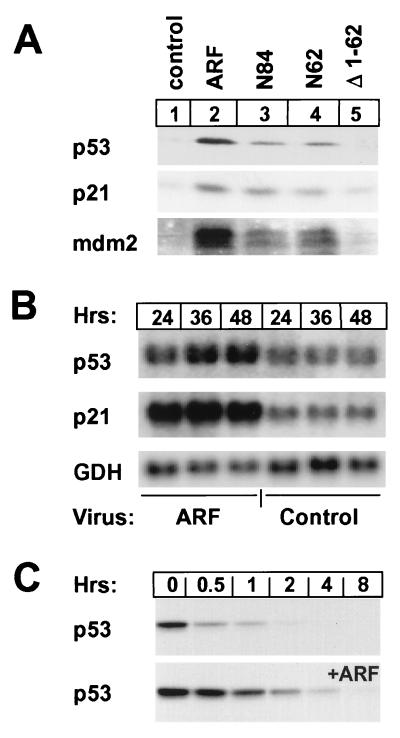
Proliferating ARF-null MEFs arrest in both the G1 and G2 phase of the cell cycle when infected with an ARF-containing retrovirus (2, 34), which induces the cyclin-dependent kinase inhibitor p21Cip1 in a p53-dependent manner (14). A retrovirus encoding p19ARF induced p53, p21Cip1, and mdm2 proteins in these cells (Fig. 1A, lanes 1 and 2) and in wild-type MEFs, but neither p21 nor mdm2 were induced in p53-null cells (14, 24, 25). The 62 N-terminal amino acids of mouse p19ARF are necessary and sufficient to block proliferation (34). Infection of ARF-null (or wild-type) MEFs with truncation mutants retaining the N-terminal 84 (N84) and 62 (N62) amino acids of p19ARF led to accumulation of p53, p21, and mdm2 (lanes 3 and 4), but an ARF mutant lacking amino acids 1–62 was inactive (lane 5). About 2-fold elevated levels of p53 mRNA accumulated in growth-arrested cells infected with the ARF retrovirus vs. those detected in uninfected proliferating cells (not shown) or in cells infected with a control vector (Fig. 1B), and the half-life of the p53 protein was significantly extended from 15 to ≈75 min by 48 hr after infection (Fig. 1C). ARF-infected cells expressed more p21 (Fig. 1B) and mdm2 mRNA than cells infected with the control virus, implying that increased levels of the latter proteins (Fig. 1A) resulted at least in part from new transcription.
Figure 1.
ARF stabilizes p53 and induces p53-dependent gene expression. (A) MEFs were lysed 48 hr after infection with retroviruses encoding either p19ARF, C-terminally truncated p19ARF mutants (N84, N62), or an N-terminally truncated ARF mutant (Δ1–62). Proteins were detected by direct immunoblotting using antibodies to p53, p21Cip1, and mdm2 as indicated in the left margin. (B) Northern blot analysis of RNA extracted from MEFs infected for the indicated times with p19ARF or control retrovirus vectors. Uninfected proliferating cells express levels of p53, p21Cip1, and glyceraldehyde 3-phosphate dehydrogenase (GDH) RNAs equal to those detected in cells infected with the control vector. (C) After a 36 hr infection with either a control (Top) or p19ARF retrovirus (Bottom), ARF-null MEFs were pulsed for 1 hr with [35S]methionine and chased in medium containing excess unlabeled precursor. Precipitated p53 from cells lysed at the indicated times after labeling was resolved on denaturing gels.
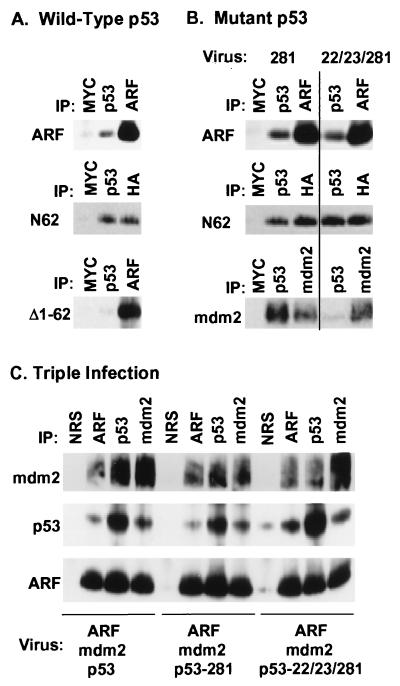
ARF can interact directly with mdm2 (24, 25), and by using Sf9 insect cells infected with baculovirus vectors encoding both proteins, binary complexes between p19ARF and mdm2 could be specifically precipitated using antisera to either (data not shown). In addition, this assay detected direct interactions between p19ARF and p53 (Fig. 2). Using wild-type p53 (Fig. 2A), antiserum to p53 coprecipitated full-length p19ARF (≈10% of input, Top) as well as the ARF N62 mutant (Middle), but not the ARF mutant lacking residues 1–62 (Bottom). Removal of the ARF C terminus potentiated its interaction with p53 (compare the amount of hemagglutinin (HA)-tagged N62 precipitated with anti-HA vs. anti-p53). A p53 point mutant (281) impaired in DNA binding interacted as well as wild-type p53 with full-length p19ARF (Fig. 2B Top) and with the N62 mutant (Fig. 2B Middle); p53 (281) also retains the capability to bind mdm2 (Fig. 2B Bottom Left). However, a p53 mutant (22/23/281) that does not stably interact with mdm2 (Fig. 2B Bottom Right) (33) retained the ability to bind ARF (Fig. 2B, top two panels). Hence, ARF can interact with p53 directly and in the absence of mdm2.
Figure 2.
Functional p19ARF binds to both mdm2 and p53 and can form ternary complexes. (A) Sf9 cells coinfected for 48 hr with baculoviruses encoding wild-type p53 and either p19ARF or the indicated p19ARF mutants were lysed and precipitated with control antibody to myc (9E10), p53 (PAb 421), affinity-purified antibody to the ARF C terminus, or anti-HA (to detect HA-tagged N62). Proteins in immune complexes separated on denaturing gels were transferred to filters and detected by immunoblotting with anti-ARF or anti-HA (for N62). (B) Similar experiments to those shown in A were performed using the indicated p53 mutants (top two panels). Sf9 cells were also coinfected with the indicated p53 mutants and mdm2 (Bottom) to document the inability of the 22/23/281 mutant to bind mdm2. Proteins precipitated with PAb421 or antibody 2A10 to mdm2 were electrophoretically resolved, transferred and blotted with 2A10. (C) Sf9 cells infected with the viruses indicated below each panel were lysed and incubated with nonimmune serum (NRS), antibodies to the ARF C terminus, PAb421 (p53), or antibody 2A10 (mdm2) as indicated at the top of each lane. Resolved proteins were blotted with antibodies to mdm2 (Top), p53 (Middle) or ARF (Bottom) as above.
One prediction is that ARF should form ternary complexes with mdm2 and p53 (22/23/281). In agreement, Sf9 cells coinfected with vectors encoding the three proteins yielded ternary complexes that were precipitated with antibodies to either (Fig. 2C). Ternary complexes can be formed between ARF, mdm2, and p53 under conditions where mdm2 serves as the “bridging” molecule (24, 25). Our data indicate that all binary complexes are possible and that ARF can similarly recruit p53 into complexes with mdm2.
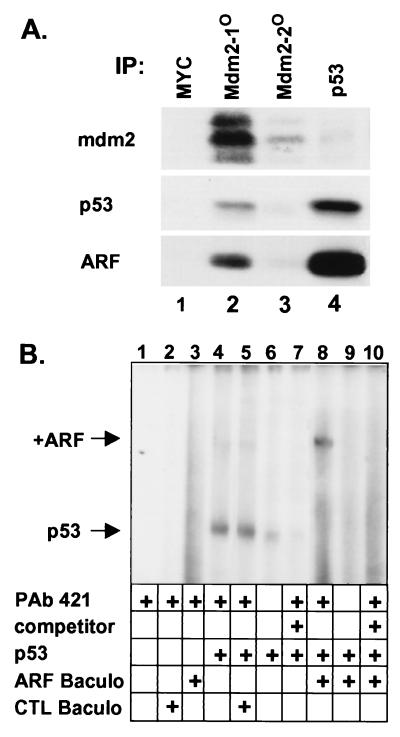
Because it is virtually impossible to assess the relative affinities of p19ARF for mdm2 and p53 in this system, we asked whether binary p19ARF-p53 complexes could be formed in ARF-null NIH 3T3 cells undergoing ARF-induced arrest (Fig. 3A). Cells infected with ARF retrovirus and lysed 48 hr after infection were precipitated sequentially with antibodies to MYC (for nonspecific binding, lane 1), twice with anti-mdm2 (lanes 2 and 3), and then with anti-p53 (lane 4). Precipitated proteins were separated and blotted with antibodies to mdm2, p19ARF, and p53. Although p19ARF and some of the induced p53 coprecipitated with anti-mdm2 (lane 2), much of the remaining p53 coprecipitated with p19ARF in binary complexes recovered from mdm2-depleted supernatants (lane 4).
Figure 3.
Direct interactions of p19ARF and p53. (A) Sequential precipitations [(IP), lanes 1–4] of lysates from NIH 3T3 cells infected with ARF virus were performed with the indicated antibodies. Precipitated proteins were separated and blotted with antibodies to mdm2, p53, and ARF. (B) EMSA was performed with an end-labeled oligonucleotide containing two consensus p53 binding sites (40). Additions to the binding reactions are indicated below the lanes and included activating antibody PAb-421, 10-fold excess cold unlabeled oligonucleotide (competitor), purified recombinant p53, and Sf9 extracts from cells infected with baculovirus vectors encoding ARF or no recombinant protein ( control, CTL). Arrows indicate positions of the p53-oligonucleotide complex and of that supershifted by ARF.
Recombinant p53 binds DNA poorly if at all, but treatment of the purified protein with an antibody (PAb 421) to a C-terminal epitope greatly enhances p53’s ability to bind radiolabeled oligonucleotides containing two consensus p53 binding sites (Fig. 3B, lanes 4 vs. 6). The complex was competed with an excess of unlabeled oligonucleotide (lane 7). Addition of extract containing p19ARF to the reaction retarded the mobility of the p53-DNA complex, (lane 8), whereas p19ARF itself did not bind the probe (lane 3). PAb 421 was required to visualize p53-DNA complexes even when p19ARF was added (lane 9), indicating that ARF did not affect p53’s ability to bind to DNA. The mdm2 protein did not bind the probe (not shown) and its addition to reactions did not affect the mobility of any of the p53-containing complexes (also see ref. 42). Therefore p19ARF can bind to p53-DNA complexes in the absence of mdm2.
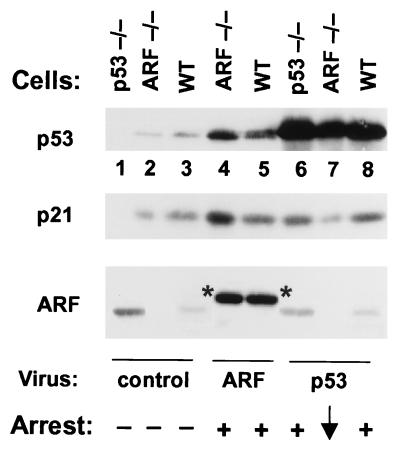
We next studied the effects of ARF and p53 retroviruses in MEFs lacking ARF or p53. MEFs derived from p53−/− embryos expressed high levels of p19ARF but reduced levels of p21Cip1 (Fig. 4, lane 1) as compared with matched, early-passage ARF-null (lane 2) or wild-type (lane 3) MEF strains. When infected with retrovirus encoding HA-tagged p19ARF, p53 and p21 were induced in both ARF-positive (lane 5 vs. 3) and ARF-null (lane 4 vs. 2) cells, and the cells underwent growth arrest. However, p53-negative cells were refractory to ARF (14, 24, 25). Infection of p53-null (lane 6) and wild-type (lane 8) MEFs with the p53 retrovirus also increased p21 expression and induced growth arrest. Reintroduction of p53 into p53-null MEFs reproducibly reduced ARF expression (lane 6 vs. 1), suggesting that either protein can regulate expression of the other. Surprisingly, supraphysiologic levels of p53 protein expression obtained in p53 virus-infected, ARF-null MEFs failed to induce p21 and had very limited effects on cell proliferation (lane 7).
Figure 4.
Induction of p21Cip1 by ARF or p53 retroviral vectors. Cells infected with vector alone (lanes 1–3), a retrovirus encoding HA-tagged p19ARF (lanes 4 and 5), or a vector encoding wild-type p53 (lanes 6–8) were lysed 48 hr after infection. Proteins separated on gels were immunoblotted for p53 (Top), p21Cip1 (Middle), and p19ARF (Bottom) as indicated at the left. Infected cells included p53-null early passage MEFs (lanes 1 and 6), wild-type MEFs (lanes 3, 5. and 8), or early passage ARF-null MEFs (lanes 2, 4, and 7). Endogenous p19ARF, elevated in p53-null cells (lane 1), is repressed after infection with p53 virus (lane 6). HA-tagged ARF (indicated by asterisks) migrates slower than the endogenous protein. Growth arrest was assayed at 48 hr by incorporation of [3H]-thymidine in replica plates.
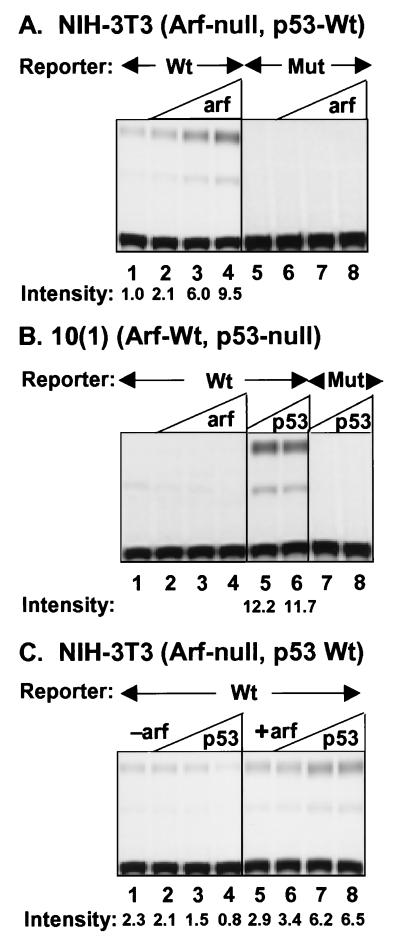
In ARF-null NIH 3T3 cells that retain functional p53, introduction of p19ARF can induce the expression of a CAT reporter gene that contains wild-type p53 binding sites in its promoter (Fig. 5A, lanes 1–4; ref. 24). Thus, ARF induces p53-dependent transactivation without enhancing the ability of p53 to bind to DNA (Fig. 3). ARF overexpression neither affected the activity of a promoter containing mutant p53 binding sites (Fig. 5A, lanes 5–8) nor induced expression of the wild-type promoter in 10(1) cells lacking functional p53 (Fig. 5B, lanes 1–4). However, cotransfection of a construct encoding wild-type p53 into p53-null 10(1) cells induced robust CAT activity (Fig. 5B, lanes 5 and 6).
Figure 5.
Transactivation by ARF and p53. NIH 3T3 or 10(1) fibroblasts were transiently transfected with wild-type PG13-CAT (WT) or mutant MG15-CAT (Mut) and increasing amounts p19ARF or p53. In A and B, the ARF plasmid inputs in lanes 2–4 and 6–8 were 1, 2, and 5 μg DNA, whereas only 10 and 100 ng of p53 plasmid were used in B (lanes 5–8). In C, cells received no ARF DNA (lanes 1–4) or 1 μg ARF plasmid (lanes 5–8) plus 1, 2, or 5 μg p53 plasmid (lanes 2–4 and 6–8, respectively). Cell lysates prepared 48 hr after transfection were analyzed for CAT activity. The mono- and diacetylated species are at the middle and top of the plate, respectively. Signal intensities for diacetylated forms computed by densitometry and indicated below the lanes were normalized to 1.0 (A, lane 1).
In agreement with data in Fig. 4, introduction of wild-type p53 (1–5 μg plasmid) into ARF-null NIH 3T3 cells was unable to induce p53 reporter gene expression, and, in fact, high levels repressed basal expression of the wild-type promoter (Fig. 5C, lanes 1–4). By contrast, p53-null, ARF-wt 10(1) cells were hypersensitive to p53, responding well to only 10 ng input plasmid DNA (Fig. 5B, lanes 5 and 6). To determine whether ARF might rescue p53 function in this setting, we transfected 1 μg of ARF expression plasmid, which was insufficient to fully activate the endogenous p53 response (Fig. 5A, lane 2), into ARF-null NIH 3T3 cells (Fig. 5C, lane 5). Cotransfection of wild-type p53 (1–5 μg plasmid) led to increased reporter gene expression (lanes 6–8). The fact that addition of ARF can restore a p53 response in NIH 3T3 cells provides direct evidence that ARF not only raises p53 levels (Fig. 1C) but also enhances its transcriptional activity through some additional mechanism.
DISCUSSION
We confirmed that p19ARF and mdm2 proteins can physically interact with one another, as well as recruit p53 into ternary complexes (24, 25). However, in both reconstituted Sf9 and ARF-infected NIH 3T3 cells, we also detected binary, mdm2-independent interactions between p19ARF and p53, which were mediated entirely by the N-terminal domain of ARF (amino acids 1–62) that alone is necessary and sufficient for its biologic activity (34). A mutant of p53 that does not interact with mdm2 could still interact with p19ARF, which in turn was able to bind mdm2. Binding of p19ARF and mdm2 also requires the N-terminal domain of ARF and the C-terminal moiety of mdm2 (amino acids 208–491) (25). Mdm2-independent interactions between p19ARF and p53 could also occur on DNA. Together, these results indicate that all binary combinations between p19ARF, mdm2, and p53 can form, and that either mdm2 or p19ARF can recruit p53 into ternary complexes.
Retroviral insertion of ARF into wild-type or ARF-null MEFs prolonged the half-life of the endogenous p53 protein, induced p53-dependent transcription of Cip1 and mdm2, and resulted in cell cycle arrest. By contrast, enforced expression of high levels of p53 protein activated transcription and growth arrest in wild-type or p53-null cells but were at best inefficient in doing so in ARF-null cells. Therefore, high levels of p53 per se are not sufficient to guarantee a response (for other examples, see refs. 43–47). To determine whether the failure of transduced p53 to function in ARF-null NIH 3T3 cells might be restored by p19ARF itself, we cotransfected low quantities of ARF retrovirus together with a p53-dependent CAT reporter plasmid and increasing concentrations of p53 DNA into NIH 3T3 cells. Under these conditions, levels of p19ARF that were insufficient to fully activate p53-dependent gene expression restored the ability of wild-type p53 to stimulate transcription. Yet, the requirement for ARF in these cells is not obligatory, because γ-irradiation of NIH 3T3 cells (or of ARF-null MEFs) induces an unimpaired p53 response (14). Therefore, in addition to stabilizing p53, p19ARF provides another activating signal. We consider several possible interpretations:
Inert p53 can be converted to an active DNA-binding form by antibodies to C-terminal p53 epitopes (43, 48), certain small peptides (43), C-terminal phosphorylation (44, 49) and acetylation (50), and by Ref-1 protein (51). Yet, p19ARF does not fulfill this function, because ARF-p53 complexes did not bind well to DNA unless an activating antibody was added. Instead, ARF might affect p53-mediated transactivation. A significant increase in transactivation by p53 can be induced in intact cells treated with low doses of UV light without a concomitant increase in the p53 level (43). Conversely, some kinase inhibitors block p53 activation without affecting its accumulation (45, 46). In vivo, modes of enhancing p53-dependent gene expression include phosphorylation of specific N-terminal serine residues, whose modification interferes with mdm2 binding (52, 53). Because p19ARF can interact with p53 on DNA, it could conceivably provide a coactivating signal of this type.
Others who first documented direct interactions between p19ARF and mdm2 suggested that p19ARF acts primarily on mdm2 rather than on p53 itself (24, 25). Binding of mdm2 to p53 accelerates its turnover (30, 31), and in agreement, mdm2 has been reported to act as a p53 E3 ligase that, together with UBC6, transfers ubiquitin to p53 and promotes its proteasomal degradation (54). Under normal circumstances, p53 levels are low and its turnover is rapid (t[1/2] ≈15 min in MEFs), but ARF overexpression, like irradiation, can significantly prolong p53’s half-life (t[1/2] ≈75 min). One scenario is that p19ARF stabilizes p53 by increasing the rate of mdm2 turnover (25), but our data indicate that mdm2 accumulates in response to p19ARF expression. Induction of mdm2 by p53 serves as a feedback mechanism to limit the p53 response (26), and p53 mutants that are defective in transactivation are stable because they do not induce mdm2 (30, 31). Regulation by mdm2 is critical in controlling p53, because disruption of the mdm2 gene in mice is lethal during early embryonic development unless p53 is also disabled (55, 56).
Nuclear localization of p53 is necessary for its transcription function (57, 58), but its degradation selectively occurs in the cytoplasm (59). In cells enforced to express p19ARF, high molecular weight species of p53 accumulated that likely represented polyubiquitinated forms (24), so p19ARF might not inhibit mdm2-mediated ubiquitination, but instead might prevent the degradation of ubiquitinated p53. An important feature of mdm2 is that it shuttles between the nucleus and the cytoplasm, and blocking its nuclear export stabilizes p53 and enhances the ability of mdm2 to block p53-mediated transcription (59). ARF localizes in discrete nuclear sites (2) together with mdm2 (24), so an attractive model is that ARF’s interactions with mdm2 and p53 prevent transport from the nucleus and thereby inhibit p53 turnover. In short, although an interdependence of p19ARF on p53 likely results from direct interactions between p53, p19ARF and mdm2, further mechanistic studies are clearly warranted.
Acknowledgments
We thank Arnold Levine for 10(1) cells, David Baltimore for 293T cells, Charles Sawyers for retroviral vectors, and Xiaoping He for baculoviruses encoding mutant p53. We gratefully acknowledge the assistance of Carol Bockhold, Esther Van de Kamp, Joseph Watson, Rose Mathew, and Zhen Lu, and the advice and support of our other laboratory colleagues. This work was supported in part by National Institutes of Health Grants CA-71907 (M.F.R.) and CA-63230 (G.Z.), Cancer Center CORE Grant CA-21765, and by ALSAC of St. Jude Children’s Research Hospital.
ABBREVIATIONS
- MEF
mouse embryo fibroblasts
- CAT
chloramphenicol acetyltransferase
- EMSA
electrophoretic mobility-shift assay
- HA
hemagglutinin
- PAb
p53 monoclonal antibody
References
- 1.Serrano M, Hannon G J, Beach D. Nature (London) 1993;366:704–707. doi: 10.1038/366704a0. [DOI] [PubMed] [Google Scholar]
- 2.Quelle D E, Zindy F, Ashmun R A, Sherr C J. Cell. 1995;83:993–1000. doi: 10.1016/0092-8674(95)90214-7. [DOI] [PubMed] [Google Scholar]
- 3.Hollstein M, Rice K, Greenblatt M S, Soussi T, Fuchs R, Sorlie T, Hovig E, Smith-Sorensen B, Montesano R, Harris C C. Nucleic Acids Res. 1994;22:3551–3555. [PMC free article] [PubMed] [Google Scholar]
- 4.Hall M, Peters G. Adv Cancer Res. 1996;68:67–108. doi: 10.1016/s0065-230x(08)60352-8. [DOI] [PubMed] [Google Scholar]
- 5.Duro D, Bernard O, Della Valle V, Berger R, Larsen C-J. Oncogene. 1995;11:21–29. [PubMed] [Google Scholar]
- 6.Mao L, Merlo A, Bedi G, Shapiro G I, Edwards C D, Rollins B J, Sidransky D. Cancer Res. 1995;55:2995–2997. [PubMed] [Google Scholar]
- 7.Stone S, Jiang P, Dayananth P, Tavtigian S V, Katcher H, Parry D, Peters G, Kamb A. Cancer Res. 1995;55:2988–2994. [PubMed] [Google Scholar]
- 8.Kato J-Y, Matsushime H, Hiebert S W, Ewen M E, Sherr C J. Genes Dev. 1993;7:331–342. doi: 10.1101/gad.7.3.331. [DOI] [PubMed] [Google Scholar]
- 9.Ewen M E, Sluss H K, Sherr C J, Matsushime H, Kato J-Y, Livingston D M. Cell. 1993;73:487–497. doi: 10.1016/0092-8674(93)90136-e. [DOI] [PubMed] [Google Scholar]
- 10.Lukas J, Müller H, Bartkova J, Spitkovsky D, Kjerulff A A, Jansen-Dürr P, Strauss M, Bartek J. J Cell Biol. 1994;125:625–638. doi: 10.1083/jcb.125.3.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lukas J, Parry D, Aagaard L, Mann D J, Bartkova J, Strauss M, Peters G, Bartek J. Nature (London) 1995;375:503–506. doi: 10.1038/375503a0. [DOI] [PubMed] [Google Scholar]
- 12.Koh J, Enders G H, Dynlacht B D, Harlow E. Nature (London) 1995;375:506–510. doi: 10.1038/375506a0. [DOI] [PubMed] [Google Scholar]
- 13.Medema R H, Herrera R E, Lam F, Weinberg R A. Proc Natl Acad Sci USA. 1995;92:6289–6293. doi: 10.1073/pnas.92.14.6289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kamijo T, Zindy F, Roussel M F, Quelle D E, Downing J R, Ashmun R A, Grosveld G, Sherr C J. Cell. 1997;91:649–659. doi: 10.1016/s0092-8674(00)80452-3. [DOI] [PubMed] [Google Scholar]
- 15.Zindy F, Quelle D E, Roussel M F, Sherr C J. Oncogene. 1997;15:203–211. doi: 10.1038/sj.onc.1201178. [DOI] [PubMed] [Google Scholar]
- 16.Todaro G J, Green H. J Cell Biol. 1963;17:299–313. doi: 10.1083/jcb.17.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harvey D M, Levine A J. Genes Dev. 1991;5:2375–2385. doi: 10.1101/gad.5.12b.2375. [DOI] [PubMed] [Google Scholar]
- 18.Serrano M, Lee H-W, Chin L, Cordon-Cardo C, Beach D, DePinho R A. Cell. 1996;85:27–37. doi: 10.1016/s0092-8674(00)81079-x. [DOI] [PubMed] [Google Scholar]
- 19.Fukusawa K, Choi T, Kuriyama R, Rulong S, Vande Woude G F. Science. 1996;271:1744–1747. doi: 10.1126/science.271.5256.1744. [DOI] [PubMed] [Google Scholar]
- 20.Jacks T, Weinberg R A. Nature (London) 1996;381:643–644. doi: 10.1038/381643a0. [DOI] [PubMed] [Google Scholar]
- 21.Levine A J. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 22.Paulovich A G, Toczyski D P, Hartwell L H. Cell. 1997;88:315–321. doi: 10.1016/s0092-8674(00)81870-x. [DOI] [PubMed] [Google Scholar]
- 23.Lanni J S, Jacks T. Mol Cell Biol. 1998;18:1055–1064. doi: 10.1128/mcb.18.2.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pomerantz J, Schreiber-Agus N, Liégeois N J, Silverman A, Alland L, Chin L, Potes J, Chen K, Orlow I, Lee H-W, et al. Cell. 1998;92:713–723. doi: 10.1016/s0092-8674(00)81400-2. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Y, Xiong Y, Yarbrough W G. Cell. 1998;92:725–734. doi: 10.1016/s0092-8674(00)81401-4. [DOI] [PubMed] [Google Scholar]
- 26.Wu X, Bayle J H, Olson D, Levine A J. Genes Dev. 1993;7:1126–1132. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- 27.Momand J, Zambetti G P, Olson D C, George D, Levine A J. Cell. 1992;69:1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- 28.Oliner J D, Pietenpol J A, Thiagalingam S, Gyuris J, Kinzler K W, Vogelstein B. Nature (London) 1993;362:857–860. doi: 10.1038/362857a0. [DOI] [PubMed] [Google Scholar]
- 29.Chen J, Marechal V, Levine A J. Mol Cell Biol. 1993;13:4107–4114. doi: 10.1128/mcb.13.7.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haupt Y, Maya R, Kazaz A, Oren M. Nature (London) 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 31.Kubbutat M H, Jones S N, Vousden K H. Nature (London) 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 32.Hinds P W, Finlay C A, Quartin R S, Baker S J, Fearon E R, Vogelstein B, Levine A J. Cell Growth Diff. 1990;1:571–580. [PubMed] [Google Scholar]
- 33.Lin J, Chen J, Elenbaas B, Levine A J. Genes Dev. 1994;8:1235–1246. doi: 10.1101/gad.8.10.1235. [DOI] [PubMed] [Google Scholar]
- 34.Quelle D E, Cheng M, Ashmun R A, Sherr C J. Proc Natl Acad Sci USA. 1997;94:3436–3440. doi: 10.1073/pnas.94.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen C, Okayama H. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tan T, Wallis J, Levine A J. J Virol. 1986;59:574–583. doi: 10.1128/jvi.59.3.574-583.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kern S E, Pietenpol J A, Thiagalingam S, Seymour A, Kinzler K W, Vogelstein B. Science. 1992;256:827–830. doi: 10.1126/science.1589764. [DOI] [PubMed] [Google Scholar]
- 38.Gorman C M, Moffat L F, Howard B H. Mol Cell Biol. 1982;2:1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zambetti G P, Bargonetti J, Walker K, Prives C, Levine A J. Genes Dev. 1992;6:1143–1152. doi: 10.1101/gad.6.7.1143. [DOI] [PubMed] [Google Scholar]
- 40.Friedlander P, Legros Y, Soussi T, Prives C. J Biol Chem. 1996;271:25468–25478. doi: 10.1074/jbc.271.41.25468. [DOI] [PubMed] [Google Scholar]
- 41.Wang E H, Friedman P N, Prives C. Cell. 1989;57:379–392. doi: 10.1016/0092-8674(89)90913-6. [DOI] [PubMed] [Google Scholar]
- 42.Zauberman A, Barak Y, Ragimov N, Levy N, Oren M. EMBO J. 1993;12:2799–2808. doi: 10.1002/j.1460-2075.1993.tb05941.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hupp T R, Sparks A, Lane D P. Cell. 1995;83:237–245. doi: 10.1016/0092-8674(95)90165-5. [DOI] [PubMed] [Google Scholar]
- 44.Hao M, Lowy A M, Kapoor M, Deffie A, Liu G, Lozano G. J Biol Chem. 1996;271:29380–29385. doi: 10.1074/jbc.271.46.29380. [DOI] [PubMed] [Google Scholar]
- 45.Chernov M V, Stark G R. Oncogene. 1997;14:2503–2510. doi: 10.1038/sj.onc.1201104. [DOI] [PubMed] [Google Scholar]
- 46.Chernov M V, Ramana C V, Adler V V, Stark G R. Proc Natl Acad Sci USA. 1998;95:2284–2289. doi: 10.1073/pnas.95.5.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Garkavtsev I, Grigorian I A, Ossovskaya V S, Chernov M V, Chumakov P M, Gudkov A V. Nature (London) 1997;391:295–298. doi: 10.1038/34675. [DOI] [PubMed] [Google Scholar]
- 48.Hupp T R, Meek D W, Midgley C A, Lane D P. Cell. 1992;71:875–886. doi: 10.1016/0092-8674(92)90562-q. [DOI] [PubMed] [Google Scholar]
- 49.Kapoor M, Lozano G. Proc Natl Acad Sci USA. 1998;95:2834–2837. doi: 10.1073/pnas.95.6.2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gu W, Roeder R G. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 51.Jayaraman L, Murthy K G K, Zhu C, Curran T, Xanthoudakis S, Prives C. Genes Dev. 1997;11:558–570. doi: 10.1101/gad.11.5.558. [DOI] [PubMed] [Google Scholar]
- 52.Shieh S-Y, Ikeda M, Taya Y, Prives P. Cell. 1997;91:325–334. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- 53.Siliciano J D, Canman C E, Taya Y, Sakaguchi K, Appella E, Kastan M B. Genes Dev. 1997;11:3471–3481. doi: 10.1101/gad.11.24.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Honda R, Tanaka H, Yasuda H. FEBS Letts. 1997;420:25–27. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- 55.Montes de Oca Luna R, Wagner D S, Lozano G. Nature (London) 1995;378:203–206. doi: 10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- 56.Jones S N, Roe A E, Donehower L A, Bradley A. Nature (London) 1995;378:206–208. doi: 10.1038/378206a0. [DOI] [PubMed] [Google Scholar]
- 57.Gannon J V, Lane D P. Nature (London) 1991;349:802–806. doi: 10.1038/349802a0. [DOI] [PubMed] [Google Scholar]
- 58.Shaulsky G, Goldfinger N, Tosky M S, Levine A J, Rotter V. Oncogene. 1991;6:2055–2065. [PubMed] [Google Scholar]
- 59.Roth J, Dobbelstein M, Freedman D, Shenk T, Levine A J. EMBO J. 1998;17:554–564. doi: 10.1093/emboj/17.2.554. [DOI] [PMC free article] [PubMed] [Google Scholar]