Abstract
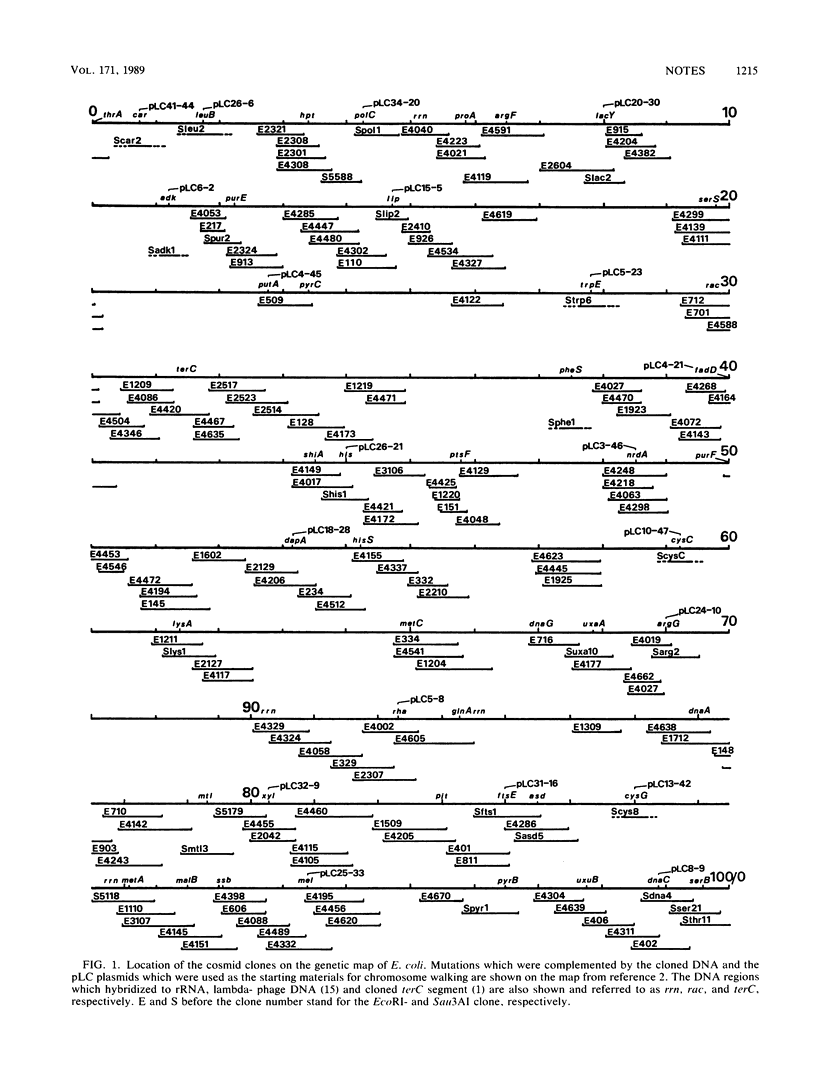
A cosmid library of the Escherichia coli K-12 W3110 chromosome was constructed in which clones were assigned to locations on the chromosome map by hybridization and genetic marker complementation tests. Approximately 70% of the genome was represented by this library. The identified clones can be maintained in the homologous system and would facilitate genetic studies of E. coli.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Asada K., Nakatani S., Takanami M. Cloning of the contiguous 165-kilobase-pair region around the terminus of Escherichia coli K-12 DNA replication. J Bacteriol. 1985 Jul;163(1):398–400. doi: 10.1128/jb.163.1.398-400.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann B. J. Linkage map of Escherichia coli K-12, edition 7. Microbiol Rev. 1983 Jun;47(2):180–230. doi: 10.1128/mr.47.2.180-230.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg C. M., Curtiss R., 3rd Transposition derivatives of an Hfr strain of Escherichia coli K-12. Genetics. 1967 Jul;56(3):503–525. doi: 10.1093/genetics/56.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitner R. M., Kuempel P. L. P1 transduction map spanning the replication terminus of Escherichia coli K12. Mol Gen Genet. 1981;184(2):208–212. doi: 10.1007/BF00272906. [DOI] [PubMed] [Google Scholar]
- Bohin J. P., Kennedy E. P. Mapping of a locus (mdoA) that affects the biosynthesis of membrane-derived oligosaccharides in Escherichia coli. J Bacteriol. 1984 Mar;157(3):956–957. doi: 10.1128/jb.157.3.956-957.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carl P. L. Escherichia coli mutants with temperature-sensitive synthesis of DNA. Mol Gen Genet. 1970;109(2):107–122. doi: 10.1007/BF00269647. [DOI] [PubMed] [Google Scholar]
- Chen P. L., Carl P. L. Genetic map location of the Escherichia coli dnaG gene. J Bacteriol. 1975 Dec;124(3):1613–1614. doi: 10.1128/jb.124.3.1613-1614.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke L., Carbon J. A colony bank containing synthetic Col El hybrid plasmids representative of the entire E. coli genome. Cell. 1976 Sep;9(1):91–99. doi: 10.1016/0092-8674(76)90055-6. [DOI] [PubMed] [Google Scholar]
- Clarke L., Carbon J. Biochemical construction and selection of hybrid plasmids containing specific segments of the Escherichia coli genome. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4361–4365. doi: 10.1073/pnas.72.11.4361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper S. Utilization of d-Methionine by Escherichia coli. J Bacteriol. 1966 Aug;92(2):328–332. doi: 10.1128/jb.92.2.328-332.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulson A., Sulston J., Brenner S., Karn J. Toward a physical map of the genome of the nematode Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7821–7825. doi: 10.1073/pnas.83.20.7821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels D. L., Blattner F. R. Mapping using gene encyclopaedias. 1987 Feb 26-Mar 4Nature. 325(6107):831–832. doi: 10.1038/325831a0. [DOI] [PubMed] [Google Scholar]
- Evans R., Seeley N. R., Kuempel P. L. Loss of rac locus DNA in merozygotes of Escherichia coli K12. Mol Gen Genet. 1979 Oct 1;175(3):245–250. doi: 10.1007/BF00397223. [DOI] [PubMed] [Google Scholar]
- Fuchs J. A., Karlström H. O. Mapping of nrdA and nrdB in Escherichia coli K-12. J Bacteriol. 1976 Dec;128(3):810–814. doi: 10.1128/jb.128.3.810-814.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glassberg J., Meyer R. R., Kornberg A. Mutant single-strand binding protein of Escherichia coli: genetic and physiological characterization. J Bacteriol. 1979 Oct;140(1):14–19. doi: 10.1128/jb.140.1.14-19.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goss T. J., Datta P. Escherichia coli K-12 mutation that inactivates biodegradative threonine dehydratase by transposon Tn5 insertion. J Bacteriol. 1984 Jun;158(3):826–831. doi: 10.1128/jb.158.3.826-831.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOWARD-FLANDERS P., SIMSON E., THERIOT L. A LOCUS THAT CONTROLS FILAMENT FORMATION AND SENSITIVITY TO RADIATION IN ESCHERICHIA COLI K-12. Genetics. 1964 Feb;49:237–246. doi: 10.1093/genetics/49.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henson J. M., Blinkowa A., Walker J. R. The Escherichia coli dnaW mutation is an allele of the adk gene. Mol Gen Genet. 1982;186(4):488–492. doi: 10.1007/BF00337953. [DOI] [PubMed] [Google Scholar]
- Herbert A. A., Guest J. R. Biochemical and genetic studies with lysine+methionine mutants of Escherichia coli: lipoic acid and alpha-ketoglutarate dehydrogenase-less mutants. J Gen Microbiol. 1968 Oct;53(3):363–381. doi: 10.1099/00221287-53-3-363. [DOI] [PubMed] [Google Scholar]
- Hill C. W., Harnish B. W. Inversions between ribosomal RNA genes of Escherichia coli. Proc Natl Acad Sci U S A. 1981 Nov;78(11):7069–7072. doi: 10.1073/pnas.78.11.7069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmeyer J., Neuhard J. Metabolism of exogenous purine bases and nucleosides by Salmonella typhimurium. J Bacteriol. 1971 Apr;106(1):14–24. doi: 10.1128/jb.106.1.14-24.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohn B., Collins J. A small cosmid for efficient cloning of large DNA fragments. Gene. 1980 Nov;11(3-4):291–298. doi: 10.1016/0378-1119(80)90069-4. [DOI] [PubMed] [Google Scholar]
- Hohn B. In vitro packaging of lambda and cosmid DNA. Methods Enzymol. 1979;68:299–309. doi: 10.1016/0076-6879(79)68021-7. [DOI] [PubMed] [Google Scholar]
- Jones-Mortimer M. C., Kornberg H. L. Genetical analysis of fructose utilization by Escherichia coli. Proc R Soc Lond B Biol Sci. 1974 Sep 17;187(1087):121–131. doi: 10.1098/rspb.1974.0066. [DOI] [PubMed] [Google Scholar]
- Kadner R. J., Winkler H. H. Isolation and characterization of mutations affecting the transport of hexose phosphates in Escherichia coli. J Bacteriol. 1973 Feb;113(2):895–900. doi: 10.1128/jb.113.2.895-900.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohara Y., Akiyama K., Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987 Jul 31;50(3):495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- Low B. Formation of merodiploids in matings with a class of Rec- recipient strains of Escherichia coli K12. Proc Natl Acad Sci U S A. 1968 May;60(1):160–167. doi: 10.1073/pnas.60.1.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low B., Gates F., Goldstein T., Söll D. Isolation and partial characterization of temperature-sensitive Escherichia coli mutants with altered leucyl- and seryl-transfer ribonucleic acid synthetases. J Bacteriol. 1971 Nov;108(2):742–750. doi: 10.1128/jb.108.2.742-750.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura Y., Caro L., Berg C. M., Hirota Y. Chromosome replication in Escherichia coli. IV. Control of chromosome replication and cell division by an integrated episome. J Mol Biol. 1971 Feb 14;55(3):441–456. doi: 10.1016/0022-2836(71)90328-7. [DOI] [PubMed] [Google Scholar]
- Oliver D. B. Identification of five new essential genes involved in the synthesis of a secreted protein in Escherichia coli. J Bacteriol. 1985 Jan;161(1):285–291. doi: 10.1128/jb.161.1.285-291.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson M. V., Dutchik J. E., Graham M. Y., Brodeur G. M., Helms C., Frank M., MacCollin M., Scheinman R., Frank T. Random-clone strategy for genomic restriction mapping in yeast. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7826–7830. doi: 10.1073/pnas.83.20.7826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overath P., Pauli G., Schairer H. U. Fatty acid degradation in Escherichia coli. An inducible acyl-CoA synthetase, the mapping of old-mutations, and the isolation of regulatory mutants. Eur J Biochem. 1969 Feb;7(4):559–574. [PubMed] [Google Scholar]
- Pittard J., Wallace B. J. Gene controlling the uptake of shikimic acid by Escherichia coli. J Bacteriol. 1966 Oct;92(4):1070–1075. doi: 10.1128/jb.92.4.1070-1075.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricard M., Hirota Y. Process of cellular division in Escherichia coli: physiological study on thermosensitive mutants defective in cell division. J Bacteriol. 1973 Oct;116(1):314–322. doi: 10.1128/jb.116.1.314-322.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert-Baudouy J. M., Portalier R. C. Mutations affectant le catabolisme du glucuronate chez Escherichia coli K12. Mol Gen Genet. 1974;131(1):31–46. doi: 10.1007/BF00269385. [DOI] [PubMed] [Google Scholar]
- Russell R. R., Pittard A. J. Mutants of Escherichia coli unable to make protein at 42 C. J Bacteriol. 1971 Nov;108(2):790–798. doi: 10.1128/jb.108.2.790-798.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw K. J., Berg C. M. Escherichia coli K-12 auxotrophs induced by insertion of the transposable element Tn5. Genetics. 1979 Jul;92(3):741–747. doi: 10.1093/genetics/92.3.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba K., Ito K., Yura T. Mutation that suppresses the protein export defect of the secY mutation and causes cold-sensitive growth of Escherichia coli. J Bacteriol. 1984 Nov;160(2):696–701. doi: 10.1128/jb.160.2.696-701.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. L., Econome J. G., Schutt A., Klco S., Cantor C. R. A physical map of the Escherichia coli K12 genome. Science. 1987 Jun 12;236(4807):1448–1453. doi: 10.1126/science.3296194. [DOI] [PubMed] [Google Scholar]
- Symington L. S., Morrison P., Kolodner R. Intramolecular recombination of linear DNA catalyzed by the Escherichia coli RecE recombination system. J Mol Biol. 1985 Dec 5;186(3):515–525. doi: 10.1016/0022-2836(85)90126-3. [DOI] [PubMed] [Google Scholar]
- Ueno-Nishio S., Backman K. C., Magasanik B. Regulation at the glnL-operator-promoter of the complex glnALG operon of Escherichia coli. J Bacteriol. 1983 Mar;153(3):1247–1251. doi: 10.1128/jb.153.3.1247-1251.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wechsler J. A., Gross J. D. Escherichia coli mutants temperature-sensitive for DNA synthesis. Mol Gen Genet. 1971;113(3):273–284. doi: 10.1007/BF00339547. [DOI] [PubMed] [Google Scholar]
- Whalen W. A., Berg C. M. Gratuitous repression of avtA in Escherichia coli and Salmonella typhimurium. J Bacteriol. 1984 May;158(2):571–574. doi: 10.1128/jb.158.2.571-574.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijsman H. J., Pafort H. C. Pleiotropic mutations in Escherichia coli conferring tolerance to glycine and sensitivity to penicillin. Mol Gen Genet. 1974;128(4):349–357. doi: 10.1007/BF00268522. [DOI] [PubMed] [Google Scholar]


