Abstract
DNA fragments −0.57, −2.2, −2.9, −5.3, and −8.4 kb in length from the upstream regulatory region of the vnd/NK-2 gene were cloned in the 5′-flanking region of a β-galactosidase (β-gal) reporter gene in the P-element pCaSpeR-AUG-β-gal, and the effects of the DNA on the pattern and time of expression of β-gal were determined in transgenic embryos. Embryos from 11 lines transformed with −8.4 kb of vnd/NK-2 regulatory DNA expressed β-gal patterns that closely resemble those of vnd/NK-2. In embryos from four lines transformed with −5.3 kb of vnd/NK-2 DNA, β-gal was found in the normal vnd/NK-2 pattern in the nerve cord but not in part of the cephalic region. β-Gal patterns in embryos from transgenic lines containing −0.57, −2.2, or −2.9 kb of vnd/NK-2 DNA did not resemble vnd/NK-2. Null vnd/NK-2 mutant embryos containing the homozygous P-element p[−8.4 to +0.34 β-gal] expressed little β-gal in contrast to siblings with a wild-type vnd/NK-2 gene. We conclude that (i) the 8.4-kb DNA fragment from the vnd/NK-2 gene contains the nucleotide sequences required to generate the normal pattern of vnd/NK-2 gene expression, sequences that may be involved in the switch between neuroblast vs. epidermoblast pathways of development, (ii) the 5′-flanking region of the vnd/NK-2 gene between −5.3 and −8.4 kb is required for vnd/NK-2 gene expression in the most dorsoanterior part of the cephalic region, and (iii) vnd/NK-2 protein is required, directly or indirectly, for maintenance of vnd/NK-2 gene expression.
Keywords: embryonic neurogenesis/epidermogenesis/gene regulation
The vnd/NK-2 homeodomain protein (1–3) initiates the neural pathway of development in part of the ventromedial nerve cord of Drosophila embryos by activating the expression of the proneural genes achaete, scute, and lethal at scute (4). A pattern of neuroectodermal cells that express the vnd/NK-2 gene is generated. Some of these cells give rise to vnd/NK-2-expressing medial neuroblasts that are the precursors of neurons that also contain vnd/NK-2 mRNA (5). Only some neuroectodermal cells that express the vnd/NK-2 gene develop into neuroblasts; in other cells, repression of the vnd/NK-2 gene, presumably by lateral inhibition, turns off the neural pathway of development. Hence, part of the information that determines a pattern of neural cells in the CNS of developing embryos resides in the nucleotide sequences that regulate the expression of the vnd/NK-2 gene.
We have cloned DNA fragments from the upstream, regulatory region of the vnd/NK-2 gene into the 5′-flanking region of an enhancerless, promoterless β-galactosidase (β-gal) reporter gene in the P-element pCaSpeR-AUG-β-gal (6), and the DNA constructs were used to generate transgenic lines of Drosophila. We found that the cis-regulatory elements required to generate the vnd/NK-2 pattern of gene expression in the CNS during embryonic development resides in −8.4 kb of DNA from the upstream region of the vnd/NK-2 gene, that the DNA between −5.3 and −8.4 kb is required for the most dorsoanterior expression of the vnd/NK-2 gene, and that vnd/NK-2 protein, directly or indirectly, is required for maintenance of vnd/NK-2 gene expression.
MATERIALS AND METHODS
Preparation of Transgenic Fly Lines Containing vnd/NK-2 Genomic DNA–P-Element Reporter Gene Constructs.
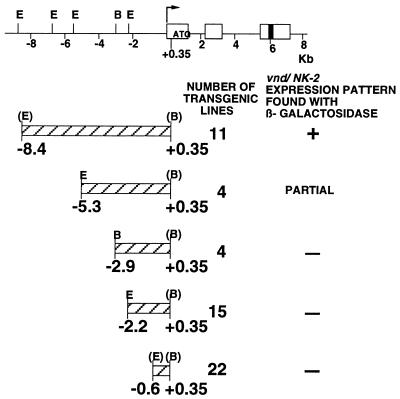
From a genomic clone of the vnd/NK-2 gene obtained by Lan Wang (National Institutes of Health, Bethesda), five P-element constructs were generated (see Fig. 2). The P-element constructs contain −8.4 to +0.35-, −5.3 to +0.35-, −2.9 to +0.35-, or −2.2 to +0.35-kb fragments of DNA from the 5′-flanking region of the vnd/NK-2 gene. These were constructed by first generating a building block, based on which larger constructs were made. The building block was made as follows. The SacII–EcoRV genomic DNA fragment corresponding to +45 to +338 bp of the vnd/NK-2 gene (Fig. 2) was ligated to two adaptors (the 5′-adaptor contained an EcoRI site, and the 3′-adaptor contained nucleotide residues +338 through +347 bp of vnd/NK-2 cDNA followed by a BamHI site) and subcloned into the EcoRI and BamHI sites of the P-element vector pCaSpeR-AUG-β-gal (6). The SacII genomic DNA fragment corresponding to −8.4 to +0.045 kb of the vnd/NK-2 gene then was inserted into this building block. The orientation was determined by restriction enzyme analysis and was confirmed by nucleotide sequencing. The −5.3 to +0.35-, −2.9 to +0.53-, and −2.2 to +0.35-kb vnd/NK-2 genomic DNA constructs were derived from the −8.4 to +0.35-kb P-element construct. The −0.6 to +0.35-kb vnd/NK-2 genomic DNA construct was generated by PCR and then subcloned into the PCaSpeR-AUG-β-gal vector.
Figure 2.
Schematic representation of the vnd/NK-2 genomic DNA and P-element constructs used to generate transformants. Exons are shown by white boxes; the black box represents the homeobox. The direction of transcription is indicated by an arrow. The position of the first methionine codon is indicated. Fragments cloned into the P-element vector are shown below the genomic map. E, EcoRI; B, BamHI. The number of transgenic lines obtained and presence of the normal vnd/NK-2 expression pattern found with β-gal are indicated.
The DNA constructs (800 μg/ml) and pπ 25.7 wc DNA (100 μg/ml) (7) were co-injected into y w embryos to generate transgenic lines (8). Each P-element insertion was mapped to a chromosome and made homozygous. Additional lines were generated by mobilization of a primary transformant by using the Δ2–3 (99B) genomic source of P-element transposase (9).
β-Gal Staining.
β-Gal activity was detected by using 5-bromo-4-chloro-3-indolylβ-d-galactoside (X-Gal) as a substrate. Embryos were dechorionated in 50% Clorox and fixed according to Hursh et al. (10) in 4% formaldehyde (EM Grade, Polysciences) in PBS (150 mM NaCl/1.7 mM KH2PO4/5 mM Na2HPO4, pH 7.2): n-heptane, 1:1. The fixed embryos were washed thoroughly with PBT (0.2% Triton X-100 in PBS) and stained with 0.2% X-Gal in staining solution [10 mM Na2HPO4/NaH2PO4/150 mM NaCl/1 mM MgCl2/3.3 mM K4Fe(CN)6/3.3 mM K3Fe(CN)6] by using procedures described in Blackman et al. (11).
β-Galactosidase protein was detected by using polyclonal rabbit antibody directed against β-gal (Cappel) and peroxidase-conjugated goat antibody directed against rabbit IgG (Jackson ImmunoResearch). The procedure of Patel et al. (12) and the DAB substrate kit (Vector Laboratories) as color-developing reagents were used.
In Situ Hybridization.
β-Gal mRNA was detected by using an anti-sense RNA probe corresponding to the 1.5-kb Xba–SacI fragment of the β-gal. Whole-mount embryo in situ hybridizations were performed according to the procedure of Tautz and Pfeifle (13). All embryos were cleared in 70% glycerol and photographed by using the high definition 3D microscope R400 (Edge Scientific Instruments, Santa Monica, CA).
Fly Stocks.
Standard Drosophila husbandry procedures were used. Different alleles of vnd/NK-2 mutants were used to generate flies with genotype vnd/NK-2/FM7c, p[ftz promoter-β-gal]; p[−8.4 to +0.35 vnd/NK-2-β-gal]/p[−8.4 to +0.35 vnd/NK-2-β-gal] by crossing vnd/NK-2/FM7c, p[ftz promoter-β-gal]; +/+ flies with a transgenic fly line containing −8.4 to +0.35 kb of DNA from the 5′-flanking region of the vnd/NK-2 gene inserted in the pCaSpeR-AUG-β-gal; the recombinant P-element is inserted in the 2nd chromosome of Drosophila. Among the vnd/NK-2 mutants tested, vnd5, vnd6, and l(1)101/FM7c were the kind gifts of F. Jimenez (Universidad Autónoma/CSIC, Madrid), and vnd19 was a gift from N. Perrimon (Harvard Medical School, Cambridge, MA).
RESULTS AND DISCUSSION
Dorsal-Ventral and Anterior-Posterior Patterning Genes Regulate the vnd/NK-2 Gene Expression.
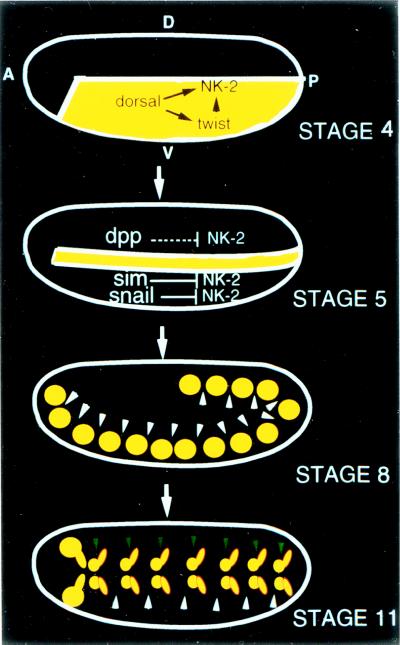
Expression of the vnd/NK-2 gene (4) initiates neural development of ventromedial neuroectodermal cells in the central nervous system of Drosophila embryos. Studies of the pattern of vnd/NK-2 gene expression in the various mutant lines reveal that the vnd/NK-2 expression is restricted to cells that give rise to the medial part of the ventral nerve cord by dorsal-ventral patterning genes (5). The vnd/NK-2 gene is activated initially in the ventral half of the embryo during late stage 4 by dorsal and twist but is repressed by snail in the mesoderm, by sim in the mesectoderm, and by dpp, mediated by an unknown repressor in the dorsal neuroectoderm (Fig. 1). The pattern of vnd/NK-2 gene expression first appears as two longitudinal stripes during stages 4–7. The stripe of neuroectodermal cells that express the vnd/NK-2 gene is converted into one cluster of vnd/NK-2- positive cells per hemisegment by periodic repression of the vnd/NK-2 gene (stages 6–8). During stages 9–11, each cluster is subdivided into two clusters of neural cells per hemisegment, suggesting a further repression of the vnd/NK-2 gene expression.
Figure 1.
Dorsal-ventral and anterior-posterior patterning genes regulate vnd/NK-2 gene expression (5). The vnd/NK-2 gene is activated initially by dorsal in the ventral half of the embryo (stage 4). Both dorsal and twist are required to activate vnd/NK-2 gene in the hindgut and posterior midgut primordia. The vnd/NK-2 gene is not expressed in the mesodermal anlage because of repression by snail, not in the mesectodermal anlage because of repression by sim, or in part of the lateral neuroectodermal and dorsal epidermal anlagen because of repression mediated by dpp. The stripe of neuroectodermal cells that express the vnd/NK-2 gene is converted into one cluster of vnd/NK-2-positive cells per hemisegment by periodic repression (indicated by white arrowheads) of the vnd/NK-2 gene (stage 8). Another kind of repressor (indicated by green arrowheads) during stages 9–11 converts each cluster into two clusters of neural cells per hemisegment. Arrows represent gene activation, and terminal bars represent gene repression. dpp indirectly mediates repression of the vnd/NK-2 gene in dorsal neuroectoderm via an unidentified repressor. A, anterior; P, posterior; D, dorsal; V, ventral.
Generation of vnd/NK-2 Genomic DNA–P-Element Constructs.
The temporal- and spatial- specific patterns of vnd/NK-2 gene expression reveal that the expression of the vnd/NK-2 gene is regulated strictly. To identify the regulatory DNA that confers the pattern of vnd/NK-2 gene expression, DNA fragments −0.6, −2.2, −2.9, −5.3, and −8.4 kb in length from the upstream regulatory region of the vnd/NK-2 gene were subcloned in the 5′-flanking region of an enhancerless, promoterless β-gal reporter gene in the P-element pCaSpeR-AUG-β-gal (Fig. 2), and the effects of the DNA inserts on the pattern and time of expression of β-gal were determined in embryos from transgenic lines of Drosophila.
Expression of vnd/NK-2 Genomic DNA-P-Element Constructs.
Embryos from 11 transgenic lines of Drosophila transformed with a P-element construct that contains −8.4 kb of regulatory DNA from the vnd/NK-2 gene expressed β-gal patterns that closely resemble the normal vnd/NK-2 patterns of gene expression (Figs. 3 and 4). Four transgenic lines of flies with P-elements containing −5.3 to +0.35 kb of vnd/NK-2 DNA inserts expressed β-gal in the normal vnd/NK-2 pattern in the ventral nerve cord but lacked the most dorsoanterior expression in the cephalic region of the embryos (Fig. 5). However, transgenic lines of flies with P-elements containing −2.9, −2.2, or −0.6 kb of vnd/NK-2 DNA inserts exhibited β-gal expression patterns in embryos that did not resemble the vnd/NK-2 pattern of gene expression. These results show that −5.3 to +0.35 kb DNA from the 5′-flanking region of the vnd/NK-2 gene contains most of the nucleotide sequences needed to activate and/or repress the vnd/NK-2 gene to generate the normal endogenous pattern of vnd/NK-2 gene expression in the ventral nerve cord and that an additional nucleotide sequence between −8.4 and −5.3 kb of vnd/NK-2 DNA is needed to generate the most dorsoanterior expression of the vnd/NK-2 gene in the cephalic region of the embryo. Partial vnd/NK-2 patterns were not detected with −2.9-kb or smaller DNA fragments from the regulatory region of the vnd/NK-2 gene.
Figure 3.
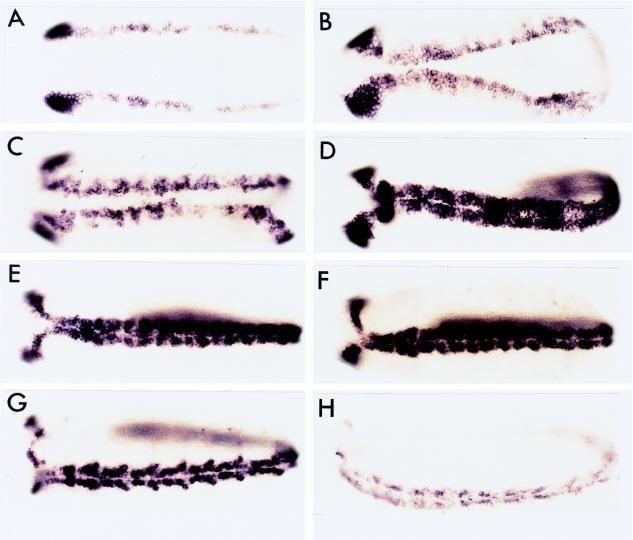
DNA (−8.4 to +0.35 kb) from the vnd/NK-2 gene confers the vnd/NK-2 pattern upon the expression of β-gal mRNA in transgenic fly lines. Embryos were stained for β-gal mRNA by using a β-gal RNA probe. No difference was detected in the pattern of expression of β-gal mRNA and vnd/NK-2 mRNA; however, during stages 5 through 7, the number of cells that expresses β-gal mRNA may be lower than those that express vnd/NK-2 mRNA. (A) Stage 5. (B) late Stage 6. (C) Stage 7. (D) Stage 8. (E) Stage 9. (F) Stage 10. (G) Stage 11. (H) Stage 12. (A–F) Ventral view. (G and H) Ventrolateral view.
Figure 4.
DNA (−8.4 to +0.35 kb) from the vnd/NK-2 gene confers the vnd/NK-2 pattern upon β-gal gene expression in transgenic fly lines. Embryos were stained for β-gal by using antibody against β-gal protein. (A) Stage 6 or 7, ventral view. (B) Stage 8, side view. (C) Stage 9, ventral view. (D) Stage 10, ventral view. (E) Stage 11, ventrolateral view. (F) Higher magnification of the embryo in E. (G) Stage 12, ventral view. (H) Higher magnification of the embryo in G. Embryos shown in the panels are all from the same transgenic fly line except the one in the D, which is from a different transgenic fly line.
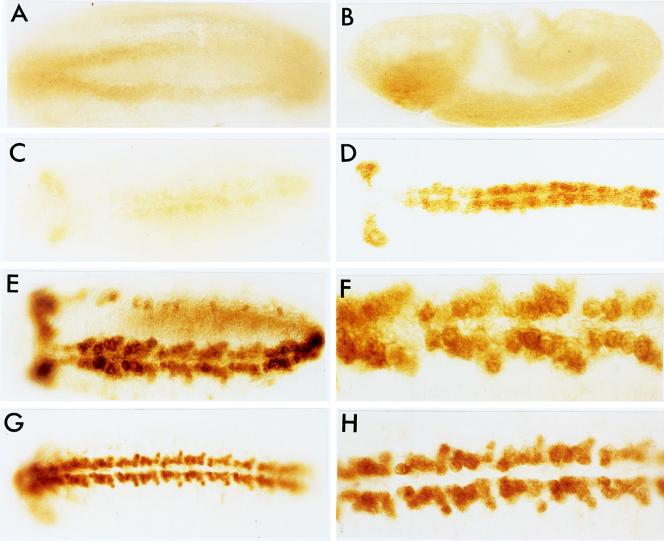
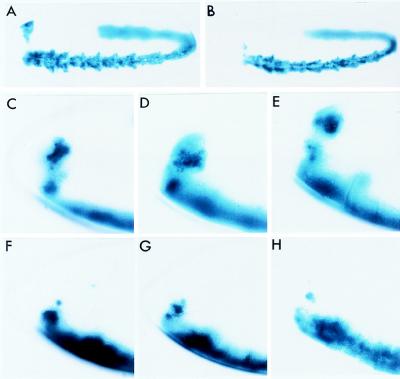
Figure 5.
DNA (−5.3 to +0.35 kb) from the vnd/NK-2 gene confers a partial vnd/NK-2 pattern upon β-gal gene expression in transgenic fly lines. Embryos were stained for β-gal activity by using X-Gal as a substrate. (Top) Ventrolateral view of late stage 11 embryos containing −8.4 to +0.35 kb (A) and −5.3 to 0.35 kb (B) DNA. (Middle and Bottom) Magnified, anterior portions of side view embryos containing −8.4 to +0.35 kb (C–E) and −5.3 to +0.35 kb (F–H) DNA at stages 11, 12, and 13 (from left to right).
Expression of the P-Element Construct p[−8.4 to +0.35 vnd/NK-2-β-gal] in vnd/NK-2 Mutant Flies.
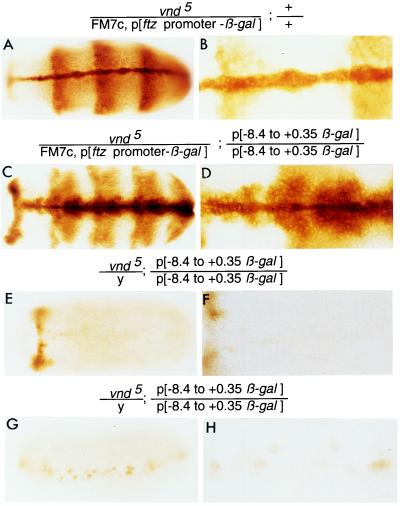
To determine whether vnd/NK-2 protein is required for expression of the vnd/NK-2 gene, the expression of the β-gal reporter gene in the P-element construct p[−8.4 to +0.35 vnd/NK-2-β-gal] was examined in the presence or absence of a functional vnd/NK-2 gene. In Fig. 6 A and B, a control embryo is shown that expresses β-gal protein in the ftz zebra pattern (and etopically in the ventral midline) due to the presence of a P-element containing the ftz zebra stripe promoter ligated to a β-gal reporter gene inserted in an FM7c balancer chromosome that contains the wild-type vnd/NK-2 gene. Hence, the ftz zebra stripe β-gal expression is a marker for the FM7c chromosome. The other chromosome 1 contains a mutant vnd5 gene. The pattern of β-gal protein shown in Fig. 6 C and D is a composite due to activation of a β-gal gene by the ftz zebra promoter inserted in chromosome 1 and expression of another β-gal reporter gene activated by −8.4 to +0.35 kb of DNA from the 5′-upstream region of the vnd/NK-2 gene ligated to a promotorless β-gal gene in homozygous P elements inserted in the second chromosome that are expressed in the wild-type vnd/NK-2 neurogenic pattern. Fig. 6 E–H show two male vnd/NK-2 embryos. In the absence of functional vnd/NK-2 protein, little β-gal protein is synthesized. These results show that activation of the β-gal reporter gene by DNA from the 5′-flanking region of the vnd/NK-2 gene depends, directly or indirectly, on functional vnd/NK-2 protein, which confirms and extends findings reported by Jimenez et al. (3). The same results were obtained with three other vnd/NK-2 mutants, vnd16, vnd19, and l(1) 101/FM7c. The results also suggest that the −8.4-kb DNA fragment from the 5′-flanking region of the vnd/NK-2 gene contains most or all of the nucleotide sequences that regulate vnd/NK-2 gene expression.
Figure 6.
vnd/NK-2 protein, directly or indirectly, activates the expression of the vnd/NK-2 gene. Expression of the β-gal reporter gene depends on functional vnd/NK-2 protein. (A and B) A female embryo with the genotype vnd/NK-2/FM7c, p[ftz promoter-β-gal]; +/+. The pattern of expression of β-gal is due to the p[ftz promoter-β-gal] insert in the FM7c balancer chromosome. (C and D) A female embryo with genotype vnd5/NK-2/FM7c, p[ftz promoter-β-gal]; p[−8.4 to +0.35 vnd/NK-2-β-gal]/p[−8.4 to +0.35 vnd/NK-2-β-gal]. The FM7c balancer chromosome contains a wild-type vnd/NK-2 gene and a DNA insert containing regulatory DNA from the ftz gene ligated to a β-gal reporter gene. The pattern of expression of β-gal protein is a composite of the vnd/NK-2 and ftz patterns of expression and etopic expression of β-gal along the ventral midline due to a positional effect on β-gal expression from the ftz regulatory DNA–β-gal reporter gene. (E–H) Male embryos with the vnd5 mutant gene and the homozygous P-element construct p[−8.4 to +0.35 β-gal] inserted in the second chromosome express little β-gal protein because mutant embryos lack wild-type functional vnd/NK-2 protein. All embryos are stage 11 embryos except the embryo in G, which is stage 12. The magnification of each embryo is higher in the right panel than in the left panel.
Nucleotide sequence analysis revealed many putative high affinity binding sites for vnd/NK-2 homeodomain protein in the −8.4-kb DNA from the 5′-flanking region of the vnd/NK-2 gene (L.-H. Wang, R. Chemelik, X. Shao, and M.N., unpublished results). However, further work is needed to determine whether the positive autoregulation of the vnd/NK-2 gene is a direct or indirect effect of vnd/NK-2 protein. Both genetic and molecular analysis have shown that maintenance of expression of some homeobox genes such as Ubx (14) or Dfd (15) depends on the protein encoded by the corresponding gene.
Effects of the vnd/NK-2 Gene on Proneural and Proepidermal Genes.
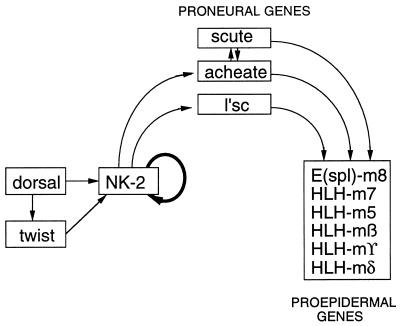
Some of the interactions of the vnd/NK-2 with other genes are summarized in Fig. 7. The vnd/NK-2 gene is activated initially in Drosophila embryos by dorsal protein and in the posterior part of the embryo by dorsal and/or twist in ventromedial neuroectodermal cells (5). Skeath et al. (4) have shown that vnd/NK-2 expression is required for activation of the proneural genes lethal at scute (l’sc), achaete, and scute. Kramatschek and Campos–Ortega (16) have shown that expression of proneural genes is required for activation of the enhancer of split complex of genes, which encode similar basic helix—loop–helix proteins whose expression initiates the epidermal pathway of development in Drosophila embryos. Hence, activation of the vnd/NK-2 gene results in activation of the neural pathway of development in ventromedial neuroectodermal cells and indirectly activates the epidermal pathway of development in these cells. Our results suggest that vnd/NK-2 protein also is required, directly or indirectly, for maintenance of vnd/NK-2 gene expression.
Figure 7.
Schematic diagram of a model for interactions of the vnd/NK-2 gene with proneural and proepidermal genes. Arrows indicate positive interactions.
All ventrolateral neuroectodermal are programmed to develop as neuroblasts, but during the course of development the neural program of development is turned off and the epidermoblast program is activated in ≈75% of the cells (17, 18). Only 25% of the cells segregate as neuroblasts (19). Probably, factors that regulate the expression of the vnd/NK-2 gene are part of the neuroblast–epidermoblast developmental switch, a process termed “lateral inhibition” (20).
Our present work indicates that the upstream −8.4 kb of vnd/NK-2 regulatory DNA contains the sequence information required to generate the normal pattern of vnd/NK-2 gene expression, as well as the sequence needed for repression of the vnd/NK-2 gene that can turn off the neural pathway of development in cells that develop as epidermoblasts. Hence, the −8.4 kb DNA from the 5′-flanking region of the vnd/NK-2 gene contains a regulatory sequence that may be involved in the developmental switch between the neuroblast and the epidermoblast pathways of development.
Acknowledgments
We thank Lan Wang for a vnd/NK-2 genomic DNA clone, C. Thummel for the P-element vector used, F. Jimenez and N. Perrimon for vnd/NK-2 mutants, Dr. Mary Whitely for providing the recombinant used to synthesize the RNA probe for β-gal mRNA, and Sven Beushausen and Brian Mozer for valuable discussions during the course of this study.
ABBREVIATION
- β-gal
β-glactosidase
References
- 1.Kim Y, Nirenberg M. Proc Natl Acad Sci USA. 1989;86:7716–7720. doi: 10.1073/pnas.86.20.7716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nirenberg M, Nakayama K, Nakayama N, Kim Y, Mellerick D, Wang L-H, Webber K, Lad R. Ann N Y Acad Sci. 1995;758:224–242. doi: 10.1111/j.1749-6632.1995.tb24830.x. [DOI] [PubMed] [Google Scholar]
- 3.Jimenez F, Martin-Morris L, Velasco L, Chu H, Sierra J, Rosen D, White K. EMBO J. 1995;14:3487–3495. doi: 10.1002/j.1460-2075.1995.tb07355.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Skeath J, Panganiban G, Carroll S. Development (Cambridge, UK) 1994;120:1517–1524. doi: 10.1242/dev.120.6.1517. [DOI] [PubMed] [Google Scholar]
- 5.Mellerick D, Nirenberg M. Dev Biol. 1995;171:306–316. doi: 10.1006/dbio.1995.1283. [DOI] [PubMed] [Google Scholar]
- 6.Thummel C, Boulet A, Lipshitz H. Gene. 1988;74:445–456. doi: 10.1016/0378-1119(88)90177-1. [DOI] [PubMed] [Google Scholar]
- 7.Karess R E, Rubin G M. Cell. 1984;38:135–146. doi: 10.1016/0092-8674(84)90534-8. [DOI] [PubMed] [Google Scholar]
- 8.Rubin G, Spradling A. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- 9.Robertson H M, Preston C R, Phillis R W, Johnson-Schlitz D M, Benz W K, Engels W R. Genetics. 1988;118:461–470. doi: 10.1093/genetics/118.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hursh D A, Padgett R W, Gelbart W M. Development (Cambridge, UK) 1993;117:1211–1222. doi: 10.1242/dev.117.4.1211. [DOI] [PubMed] [Google Scholar]
- 11.Blackman R K, Sanicola M, Raftery L A, Gillevet T, Gelbart W M. Development (Cambridge, UK) 1991;111:657–666. doi: 10.1242/dev.111.3.657. [DOI] [PubMed] [Google Scholar]
- 12.Patel N H, Schafer B, Goodman C S, Holmgren R. Genes Dev. 1989;3:890–904. doi: 10.1101/gad.3.6.890. [DOI] [PubMed] [Google Scholar]
- 13.Tautz D, Pfeifle C. Chromosoma. 1989;98:81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- 14.Bienz M, Tremml G. Nature (London) 1988;333:576– 578. doi: 10.1038/333576a0. [DOI] [PubMed] [Google Scholar]
- 15.Kuziora M A, McGinnis W. Cell. 1988;55:477–485. doi: 10.1016/0092-8674(88)90034-7. [DOI] [PubMed] [Google Scholar]
- 16.Kramatschek B, Campos-Ortega J A. Development (Cambridge, UK) 1994;120:815–826. doi: 10.1242/dev.120.4.815. [DOI] [PubMed] [Google Scholar]
- 17.Jimenez F, Campos-Ortega J A. J Neurogenet. 1987;4:179–200. [PubMed] [Google Scholar]
- 18.Jimenez F, Campos-Ortega J A. Neuron. 1990;5:81–89. doi: 10.1016/0896-6273(90)90036-f. [DOI] [PubMed] [Google Scholar]
- 19.Hartenstein V, Campos-Ortega J A. Wilhelm Roux’s Arch Dev Biol. 1984;193:308–325. doi: 10.1007/BF00848159. [DOI] [PubMed] [Google Scholar]
- 20.Artavanis-Tsakonas S, Delidakis C, Fehon R G. Annu Rev Cell Biol. 1991;7:427–452. doi: 10.1146/annurev.cb.07.110191.002235. [DOI] [PubMed] [Google Scholar]