Abstract
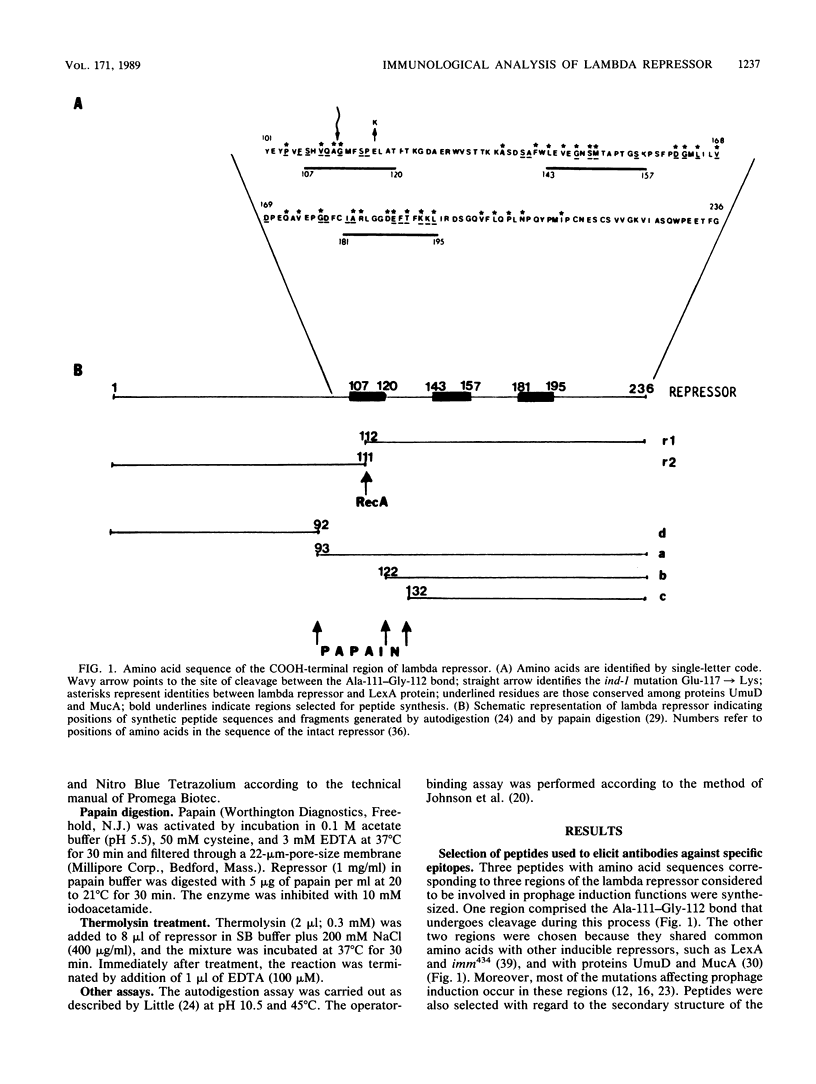
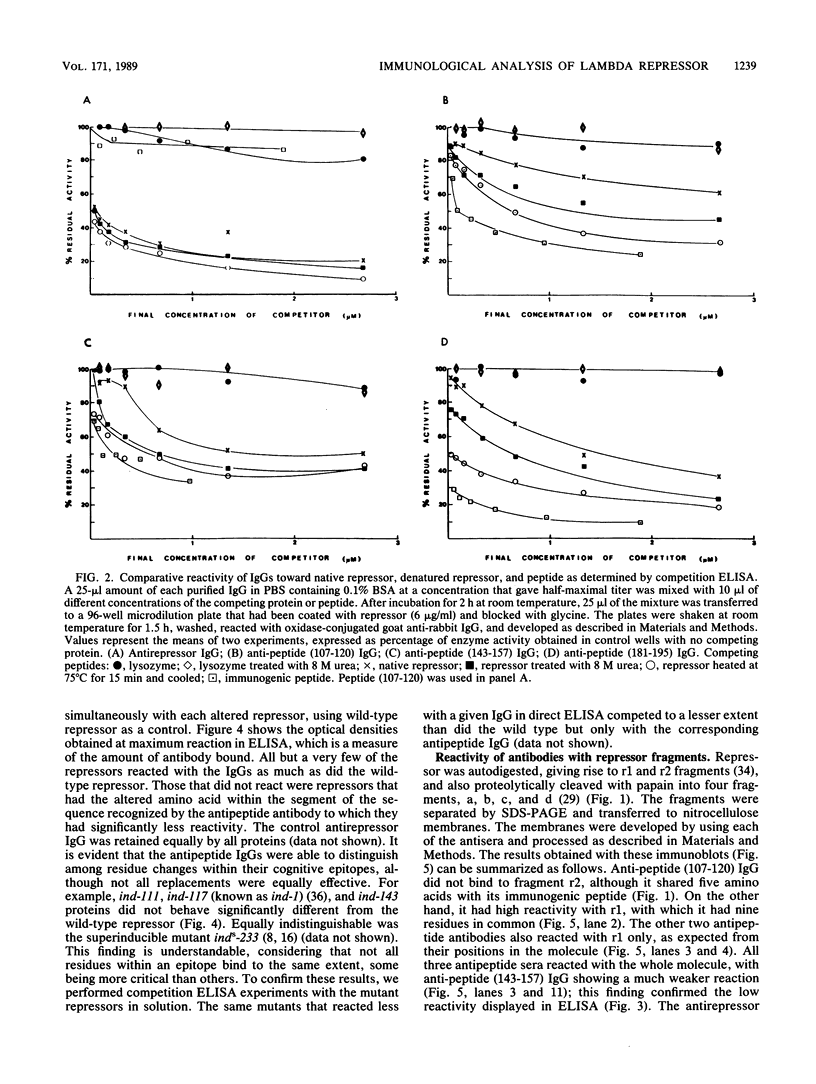
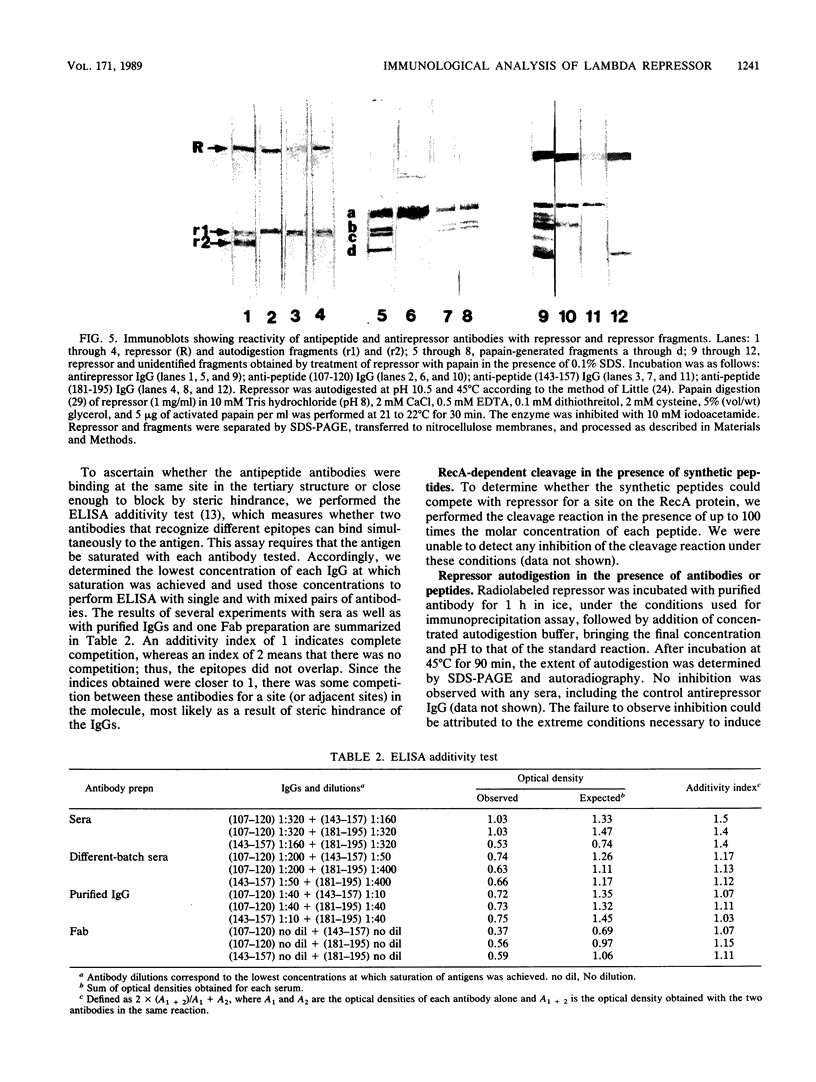
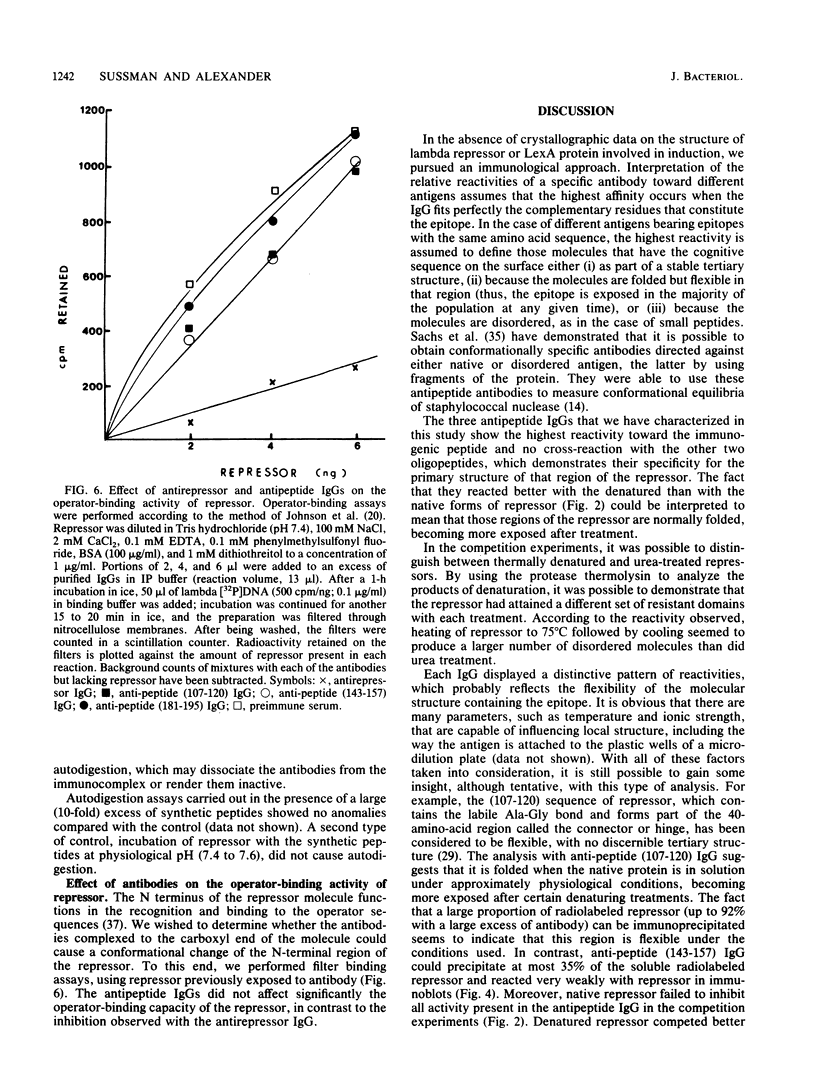
To analyze lambda repressor function and structure, antibodies were generated with synthetic peptides corresponding to sequences believed to be involved in prophage induction. These site-directed antibodies seemed to recognize preferentially the primary sequence of repressor because they reacted better in competition experiments with the oligopeptide and with the partially denatured forms of repressor than with the native molecules. This information, together with the characteristic ability of the antibodies to immunoprecipitate or react with repressor in immunoblots, allowed us to infer some conformational properties of the specific regions that the antibodies recognized. The antibodies reacted less with some mutant repressors that had a single amino acid substitution within the cognitive sequences. RecA-catalyzed cleavage of repressor was inhibited to different extents in relation to the proportion of repressor that each antipeptide immunoglobulin G (IgG) was able to immunoprecipitate. The antipeptide IgGs did not affect specific binding of repressor to operator DNA, whereas the antirepressor IgG was inhibitory. The three different IgGs competed for binding to repressor in an enzyme-linked immunosorbent assay additivity test, which suggested that the three regions of conserved amino acids are probably located on the same side of the carboxyl domain of repressor and possibly close together in the tertiary structure.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alexander H., Johnson D. A., Rosen J., Jerabek L., Green N., Weissman I. L., Lerner R. A. Mimicking the alloantigenicity of proteins with chemically synthesized peptides differing in single amino acids. Nature. 1983 Dec 15;306(5944):697–699. doi: 10.1038/306697a0. [DOI] [PubMed] [Google Scholar]
- Amann E., Brosius J., Ptashne M. Vectors bearing a hybrid trp-lac promoter useful for regulated expression of cloned genes in Escherichia coli. Gene. 1983 Nov;25(2-3):167–178. doi: 10.1016/0378-1119(83)90222-6. [DOI] [PubMed] [Google Scholar]
- Baluch J., Chase J. W., Sussman R. Synthesis of recA protein and induction of bacteriophage lambda in single-strand deoxyribonucleic acid-binding protein mutants of Escherichia coli. J Bacteriol. 1980 Nov;144(2):489–498. doi: 10.1128/jb.144.2.489-498.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baluch J., Sussman R. Correlation between UV dose requirement for lambda bacteriophage induction and lambda repressor concentration. J Virol. 1978 Jun;26(3):595–602. doi: 10.1128/jvi.26.3.595-602.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittle J. L., Houghten R. A., Alexander H., Shinnick T. M., Sutcliffe J. G., Lerner R. A., Rowlands D. J., Brown F. Protection against foot-and-mouth disease by immunization with a chemically synthesized peptide predicted from the viral nucleotide sequence. Nature. 1982 Jul 1;298(5869):30–33. doi: 10.1038/298030a0. [DOI] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol Relat Areas Mol Biol. 1978;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- Cohen S. P., Resnick J., Sussman R. Interaction of single-strand binding protein and RecA protein at the single-stranded DNA site. J Mol Biol. 1983 Jul 15;167(4):901–909. doi: 10.1016/s0022-2836(83)80119-3. [DOI] [PubMed] [Google Scholar]
- Cohen S., Knoll B. J., Little J. W., Mount D. W. Preferential cleavage of phage lambda repressor monomers by recA protease. Nature. 1981 Nov 12;294(5837):182–184. doi: 10.1038/294182a0. [DOI] [PubMed] [Google Scholar]
- Craig N. L., Roberts J. W. E. coli recA protein-directed cleavage of phage lambda repressor requires polynucleotide. Nature. 1980 Jan 3;283(5742):26–30. doi: 10.1038/283026a0. [DOI] [PubMed] [Google Scholar]
- De Anda J., Poteete A. R., Sauer R. T. P22 c2 repressor. Domain structure and function. J Biol Chem. 1983 Sep 10;258(17):10536–10542. [PubMed] [Google Scholar]
- Friguet B., Djavadi-Ohaniance L., Pages J., Bussard A., Goldberg M. A convenient enzyme-linked immunosorbent assay for testing whether monoclonal antibodies recognize the same antigenic site. Application to hybridomas specific for the beta 2-subunit of Escherichia coli tryptophan synthase. J Immunol Methods. 1983 Jun 10;60(3):351–358. doi: 10.1016/0022-1759(83)90292-2. [DOI] [PubMed] [Google Scholar]
- Furie B., Schechter A. N., Sachs D. H., Anfinsen C. B. An immunological approach to the conformational equilibrium of staphylococcal nuclease. J Mol Biol. 1975 Mar 15;92(4):497–506. doi: 10.1016/0022-2836(75)90305-8. [DOI] [PubMed] [Google Scholar]
- Gentry L. E., Rohrschneider L. R., Casnellie J. E., Krebs E. G. Antibodies to a defined region of pp60src neutralize the tyrosine-specific kinase activity. J Biol Chem. 1983 Sep 25;258(18):11219–11228. [PubMed] [Google Scholar]
- Gimble F. S., Sauer R. T. Lambda repressor inactivation: properties of purified ind- proteins in the autodigestion and RecA-mediated cleavage reactions. J Mol Biol. 1986 Nov 5;192(1):39–47. doi: 10.1016/0022-2836(86)90462-6. [DOI] [PubMed] [Google Scholar]
- Gimble F. S., Sauer R. T. Mutations in bacteriophage lambda repressor that prevent RecA-mediated cleavage. J Bacteriol. 1985 Apr;162(1):147–154. doi: 10.1128/jb.162.1.147-154.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopp T. P., Woods K. R. Prediction of protein antigenic determinants from amino acid sequences. Proc Natl Acad Sci U S A. 1981 Jun;78(6):3824–3828. doi: 10.1073/pnas.78.6.3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horii T., Ogawa T., Nakatani T., Hase T., Matsubara H., Ogawa H. Regulation of SOS functions: purification of E. coli LexA protein and determination of its specific site cleaved by the RecA protein. Cell. 1981 Dec;27(3 Pt 2):515–522. doi: 10.1016/0092-8674(81)90393-7. [DOI] [PubMed] [Google Scholar]
- Johnson A. D., Pabo C. O., Sauer R. T. Bacteriophage lambda repressor and cro protein: interactions with operator DNA. Methods Enzymol. 1980;65(1):839–856. doi: 10.1016/s0076-6879(80)65078-2. [DOI] [PubMed] [Google Scholar]
- Lerner R. A. Antibodies of predetermined specificity in biology and medicine. Adv Immunol. 1984;36:1–44. doi: 10.1016/s0065-2776(08)60898-6. [DOI] [PubMed] [Google Scholar]
- Lerner R. A., Green N., Alexander H., Liu F. T., Sutcliffe J. G., Shinnick T. M. Chemically synthesized peptides predicted from the nucleotide sequence of the hepatitis B virus genome elicit antibodies reactive with the native envelope protein of Dane particles. Proc Natl Acad Sci U S A. 1981 Jun;78(6):3403–3407. doi: 10.1073/pnas.78.6.3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieb M. A fine structure map of spontaneous and induced mutations in the lambda repressor gene, including insertions of IS elements. Mol Gen Genet. 1981;184(3):364–371. doi: 10.1007/BF00352506. [DOI] [PubMed] [Google Scholar]
- Little J. W. Autodigestion of lexA and phage lambda repressors. Proc Natl Acad Sci U S A. 1984 Mar;81(5):1375–1379. doi: 10.1073/pnas.81.5.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little J. W., Hill S. A. Deletions within a hinge region of a specific DNA-binding protein. Proc Natl Acad Sci U S A. 1985 Apr;82(8):2301–2305. doi: 10.1073/pnas.82.8.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little J. W., Mount D. W. The SOS regulatory system of Escherichia coli. Cell. 1982 May;29(1):11–22. doi: 10.1016/0092-8674(82)90085-x. [DOI] [PubMed] [Google Scholar]
- Nohmi T., Battista J. R., Dodson L. A., Walker G. C. RecA-mediated cleavage activates UmuD for mutagenesis: mechanistic relationship between transcriptional derepression and posttranslational activation. Proc Natl Acad Sci U S A. 1988 Mar;85(6):1816–1820. doi: 10.1073/pnas.85.6.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norrby E., Mufson M. A., Alexander H., Houghten R. A., Lerner R. A. Site-directed serology with synthetic peptides representing the large glycoprotein G of respiratory syncytial virus. Proc Natl Acad Sci U S A. 1987 Sep;84(18):6572–6576. doi: 10.1073/pnas.84.18.6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T., Sturtevant J. M., Ptashne M. The lambda repressor contains two domains. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1608–1612. doi: 10.1073/pnas.76.4.1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry K. L., Elledge S. J., Mitchell B. B., Marsh L., Walker G. C. umuDC and mucAB operons whose products are required for UV light- and chemical-induced mutagenesis: UmuD, MucA, and LexA proteins share homology. Proc Natl Acad Sci U S A. 1985 Jul;82(13):4331–4335. doi: 10.1073/pnas.82.13.4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phizicky E. M., Roberts J. W. Kinetics of RecA protein-directed inactivation of repressors of phage lambda and phage P22. J Mol Biol. 1980 May 25;139(3):319–328. doi: 10.1016/0022-2836(80)90133-3. [DOI] [PubMed] [Google Scholar]
- Radman M. SOS repair hypothesis: phenomenology of an inducible DNA repair which is accompanied by mutagenesis. Basic Life Sci. 1975;5A:355–367. doi: 10.1007/978-1-4684-2895-7_48. [DOI] [PubMed] [Google Scholar]
- Resnick J., Sussman R. Escherichia coli single-strand DNA binding protein from wild type and lexC113 mutant affects in vitro proteolytic cleavage of phage lambda repressor. Proc Natl Acad Sci U S A. 1982 May;79(9):2832–2835. doi: 10.1073/pnas.79.9.2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts J. W., Roberts C. W. Proteolytic cleavage of bacteriophage lambda repressor in induction. Proc Natl Acad Sci U S A. 1975 Jan;72(1):147–151. doi: 10.1073/pnas.72.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs D. H., Schechter A. N., Eastlake A., Anfinsen C. B. An immunologic approach to the conformational equilibria of polypeptides. Proc Natl Acad Sci U S A. 1972 Dec;69(12):3790–3794. doi: 10.1073/pnas.69.12.3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer R. T. DNA sequence of the bacteriophage gama cI gene. Nature. 1978 Nov 16;276(5685):301–302. doi: 10.1038/276301a0. [DOI] [PubMed] [Google Scholar]
- Sauer R. T., Pabo C. O., Meyer B. J., Ptashne M., Backman K. C. Regulatory functions of the lambda repressor reside in the amino-terminal domain. Nature. 1979 May 31;279(5712):396–400. doi: 10.1038/279396a0. [DOI] [PubMed] [Google Scholar]
- Sauer R. T., Ross M. J., Ptashne M. Cleavage of the lambda and P22 repressors by recA protein. J Biol Chem. 1982 Apr 25;257(8):4458–4462. [PubMed] [Google Scholar]
- Sauer R. T., Yocum R. R., Doolittle R. F., Lewis M., Pabo C. O. Homology among DNA-binding proteins suggests use of a conserved super-secondary structure. Nature. 1982 Jul 29;298(5873):447–451. doi: 10.1038/298447a0. [DOI] [PubMed] [Google Scholar]
- Shinagawa H., Iwasaki H., Kato T., Nakata A. RecA protein-dependent cleavage of UmuD protein and SOS mutagenesis. Proc Natl Acad Sci U S A. 1988 Mar;85(6):1806–1810. doi: 10.1073/pnas.85.6.1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slilaty S. N., Little J. W. Lysine-156 and serine-119 are required for LexA repressor cleavage: a possible mechanism. Proc Natl Acad Sci U S A. 1987 Jun;84(12):3987–3991. doi: 10.1073/pnas.84.12.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slilaty S. N., Rupley J. A., Little J. W. Intramolecular cleavage of LexA and phage lambda repressors: dependence of kinetics on repressor concentration, pH, temperature, and solvent. Biochemistry. 1986 Nov 4;25(22):6866–6875. doi: 10.1021/bi00370a020. [DOI] [PubMed] [Google Scholar]
- Susskind M. M., Botstein D. Mechanism of action of Salmonella phage P22 antirepressor. J Mol Biol. 1975 Oct 25;98(2):413–424. doi: 10.1016/s0022-2836(75)80127-6. [DOI] [PubMed] [Google Scholar]
- Sussman R., Resnick J., Calame K., Baluch J. Interaction of bacteriophage lambda repressor with nonoperator DNA containing single-strand gaps. Proc Natl Acad Sci U S A. 1978 Dec;75(12):5817–5821. doi: 10.1073/pnas.75.12.5817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlin B. E., Clark A. J. Overproduction of the Escherichia coli recA protein without stimulation of its proteolytic activity. J Bacteriol. 1981 Oct;148(1):386–390. doi: 10.1128/jb.148.1.386-390.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vales L. D., Chase J. W., Murphy J. B. Effect of ssbA1 and lexC113 mutations on lambda prophage induction, bacteriophage growth, and cell survival. J Bacteriol. 1980 Aug;143(2):887–896. doi: 10.1128/jb.143.2.887-896.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker G. C. Mutagenesis and inducible responses to deoxyribonucleic acid damage in Escherichia coli. Microbiol Rev. 1984 Mar;48(1):60–93. doi: 10.1128/mr.48.1.60-93.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstock G. M., McEntee K., Lehman I. R. ATP-dependent renaturation of DNA catalyzed by the recA protein of Escherichia coli. Proc Natl Acad Sci U S A. 1979 Jan;76(1):126–130. doi: 10.1073/pnas.76.1.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkin E. M. Ultraviolet mutagenesis and inducible DNA repair in Escherichia coli. Bacteriol Rev. 1976 Dec;40(4):869–907. doi: 10.1128/br.40.4.869-907.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]




